A multipurpose panel of microhaplotypes for use with STR markers in casework
- PMID: 35696960
- PMCID: PMC11071123
- DOI: 10.1016/j.fsigen.2022.102729
A multipurpose panel of microhaplotypes for use with STR markers in casework
Abstract
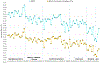
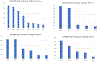
A small panel of highly informative loci that can be genotyped on the same equipment as the standard CODIS short tandem repeat (STR) markers has strong potential for application in forensic casework. Single nucleotide polymorphisms (SNPs) can be typed by a couple of methods on capillary electrophoresis (CE) machines and on sequencers, but the amount of information relative to the laboratory effort has hindered use of SNPs in actual casework. Insertion-deletion markers (InDels) suffer from similar problems. Microhaplotypes (MHs) are much more informative per locus but have similar technical difficulties unless they are typed by massively parallel sequencing (MPS). As forensic labs are acquiring sequencing machines, MHs become more likely to be used in casework, especially if multiplexed with STRs. Here we present the details of a multipurpose panel of 24 MHs with the highest effective number of alleles (Ae) from previous work. An augmented STR panel of 24 loci (20 CODIS markers plus four commonly typed STRs) is also considered. The Ae and ancestry informativeness (In) distributions of these two datasets are compared. The MH panel is shown to have better individualization and population distinction than the augmented CODIS STRs. We note that the 24 MHs should be better for mixture analyses than the STRs. Finally, we suggest that a commercial kit including both the standard CODIS markers and this set of 24 MH would greatly improve the discrimination power over that of current commercial assays.
Keywords: Ancestry; CODIS; Microhaplotype; Population genetics; SNP; STR.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
None.
Figures



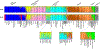


References
-
- Kidd KK, Pakstis AJ, Speed WC, Lagace R, Chang J, Wootton S, Ihuegbu N, Microhaplotype loci are a powerful new type of forensic marker, Forensic Sci. Int. Genet. Suppl. Ser. 4 (2013) e123–e124.
-
- Phillips C, McNevin D, Kidd KK, Lagace R, Wootton S, de la Puente M, Freire-Aradas A, Mosquera-Miguel A, Eduardoff M, Gross T, Dagostino L, Power D, Olson S, Hashiyada M, Oz C, Parson W, Schneider PM, Lareu MV, Daniel R, MAPlex – a massively parallel sequencing ancestry analysis multiplex for Asia-Pacific populations, Forensic Sci. Int. Genet. 42 (2019) 213–226, 10.1016/j.fsigen.2019.06.022. - DOI - PubMed
-
- de la Puente M, Phillips C, Xavier C, Amigo J, Carracedo A, Parson W, Lareu MV, Building a custom large-scale panel of novel microhaplotypes for forensic identification using MiSeq and Ion S5 massively parallel sequencing systems, Forensic Sci. Int. Genet. 45 (2020), 102213, 10.1016/j.fsigen.2019.102213. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

