Optimization of an in Silico Protocol Using Probe Permeabilities to Identify Membrane Pan-Assay Interference Compounds
- PMID: 35697029
- PMCID: PMC9770580
- DOI: 10.1021/acs.jcim.2c00372
Optimization of an in Silico Protocol Using Probe Permeabilities to Identify Membrane Pan-Assay Interference Compounds
Abstract
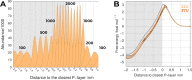
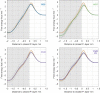
Membrane pan-assay interference compounds (PAINS) are a class of molecules that interact nonspecifically with lipid bilayers and alter their physicochemical properties. An early identification of these compounds avoids chasing false leads and the needless waste of time and resources in drug discovery campaigns. In this work, we optimized an in silico protocol on the basis of umbrella sampling (US)/molecular dynamics (MD) simulations to discriminate between compounds with different membrane PAINS behavior. We showed that the method is quite sensitive to membrane thickness fluctuations, which was mitigated by changing the US reference position to the phosphate atoms of the closest interacting monolayer. The computational efficiency was improved further by decreasing the number of umbrellas and adjusting their strength and position in our US scheme. The inhomogeneous solubility-diffusion model (ISDM) used to calculate the membrane permeability coefficients confirmed that resveratrol and curcumin have distinct membrane PAINS characteristics and indicated a misclassification of nothofagin in a previous work. Overall, we have presented here a promising in silico protocol that can be adopted as a future reference method to identify membrane PAINS.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Identification of Pan-Assay INterference compoundS (PAINS) Using an MD-Based Protocol.Methods Mol Biol. 2021;2315:263-271. doi: 10.1007/978-1-0716-1468-6_15. Methods Mol Biol. 2021. PMID: 34302681
-
In Silico Prediction of Permeability Coefficients.Methods Mol Biol. 2021;2315:243-261. doi: 10.1007/978-1-0716-1468-6_14. Methods Mol Biol. 2021. PMID: 34302680
-
Molecular dynamics simulations of ethanol permeation through single and double-lipid bilayers.J Chem Phys. 2020 Sep 28;153(12):125101. doi: 10.1063/5.0013430. J Chem Phys. 2020. PMID: 33003717 Free PMC article.
-
Molecular simulation of nonfacilitated membrane permeation.Biochim Biophys Acta. 2016 Jul;1858(7 Pt B):1672-87. doi: 10.1016/j.bbamem.2015.12.014. Epub 2015 Dec 17. Biochim Biophys Acta. 2016. PMID: 26706099 Review.
-
Benchmarking the mechanisms of frequent hitters: limitation of PAINS alerts.Drug Discov Today. 2021 Jun;26(6):1353-1358. doi: 10.1016/j.drudis.2021.02.003. Epub 2021 Feb 10. Drug Discov Today. 2021. PMID: 33581116 Review.
Cited by
-
Screening for bilayer-active and likely cytotoxic molecules reveals bilayer-mediated regulation of cell function.J Gen Physiol. 2023 Apr 3;155(4):e202213247. doi: 10.1085/jgp.202213247. Epub 2023 Feb 10. J Gen Physiol. 2023. PMID: 36763053 Free PMC article.
-
Membrane manipulation by free fatty acids improves microbial plant polyphenol synthesis.Nat Commun. 2023 Sep 12;14(1):5619. doi: 10.1038/s41467-023-40947-x. Nat Commun. 2023. PMID: 37699874 Free PMC article.
-
Recent advances on molecular dynamics-based techniques to address drug membrane permeability with atomistic detail.BBA Adv. 2023 Aug 16;4:100099. doi: 10.1016/j.bbadva.2023.100099. eCollection 2023. BBA Adv. 2023. PMID: 37675199 Free PMC article.
-
Preliminary insight on diarylpentanoids as potential antimalarials: In silico, in vitro pLDH and in vivo zebrafish toxicity assessment.Heliyon. 2024 Mar 7;10(5):e27462. doi: 10.1016/j.heliyon.2024.e27462. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38495201 Free PMC article.
-
Unraveling the Aquaporin-3 Inhibitory Effect of Rottlerin by Experimental and Computational Approaches.Int J Mol Sci. 2023 Mar 22;24(6):6004. doi: 10.3390/ijms24066004. Int J Mol Sci. 2023. PMID: 36983077 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

