Gene expression signatures of individual ductal carcinoma in situ lesions identify processes and biomarkers associated with progression towards invasive ductal carcinoma
- PMID: 35697697
- PMCID: PMC9192778
- DOI: 10.1038/s41467-022-30573-4
Gene expression signatures of individual ductal carcinoma in situ lesions identify processes and biomarkers associated with progression towards invasive ductal carcinoma
Abstract
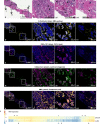
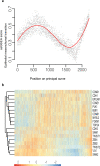
Ductal carcinoma in situ (DCIS) is considered a non-invasive precursor to breast cancer, and although associated with an increased risk of developing invasive disease, many women with DCIS will never progress beyond their in situ diagnosis. The path from normal duct to invasive ductal carcinoma (IDC) is not well understood, and efforts to do so are hampered by the substantial heterogeneity that exists between patients, and even within patients. Here we show gene expression analysis from > 2,000 individually micro-dissected ductal lesions representing 145 patients. Combining all samples into one continuous trajectory we show there is a progressive loss in basal layer integrity heading towards IDC, coupled with two epithelial to mesenchymal transitions, one early and a second coinciding with the convergence of DCIS and IDC expression profiles. We identify early processes and potential biomarkers, including CAMK2N1, MNX1, ADCY5, HOXC11 and ANKRD22, whose reduced expression is associated with the progression of DCIS to invasive breast cancer.
© 2022. The Author(s).
Conflict of interest statement
The University of Cambridge has filed a patent concerning markers identified in this study. Patent title: Biomarkers, GB priority patent application no: 2118312.4. Authors currently linked to the Patent application are C.R., G.H., J.X. and S.B. The remaining authors declare no other competing interests are present.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

