Three-nucleotide periodicity of nucleotide diversity in a population enables the identification of open reading frames
- PMID: 35698834
- PMCID: PMC9294425
- DOI: 10.1093/bib/bbac210
Three-nucleotide periodicity of nucleotide diversity in a population enables the identification of open reading frames
Abstract
Accurate prediction of open reading frames (ORFs) is important for studying and using genome sequences. Ribosomes move along mRNA strands with a step of three nucleotides and datasets carrying this information can be used to predict ORFs. The ribosome-protected footprints (RPFs) feature a significant 3-nt periodicity on mRNAs and are powerful in predicting translating ORFs, including small ORFs (sORFs), but the application of RPFs is limited because they are too short to be accurately mapped in complex genomes. In this study, we found a significant 3-nt periodicity in the datasets of populational genomic variants in coding sequences, in which the nucleotide diversity increases every three nucleotides. We suggest that this feature can be used to predict ORFs and develop the Python package 'OrfPP', which recovers ~83% of the annotated ORFs in the tested genomes on average, independent of the population sizes and the complexity of the genomes. The novel ORFs, including sORFs, identified from single-nucleotide polymorphisms are supported by protein mass spectrometry evidence comparable to that of the annotated ORFs. The application of OrfPP to tetraploid cotton and hexaploid wheat genomes successfully identified 76.17% and 87.43% of the annotated ORFs in the genomes, respectively, as well as 4704 sORFs, including 1182 upstream and 2110 downstream ORFs in cotton and 5025 sORFs, including 232 upstream and 234 downstream ORFs in wheat. Overall, we propose an alternative and supplementary approach for ORF prediction that can extend the studies of sORFs to more complex genomes.
Keywords: SNPs; open reading frame; polyploidy genome; population; sORF.
© The Author(s) 2022. Published by Oxford University Press.
Figures
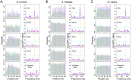


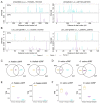



Similar articles
-
Identification of short open reading frames in plant genomes.Front Plant Sci. 2023 Feb 15;14:1094715. doi: 10.3389/fpls.2023.1094715. eCollection 2023. Front Plant Sci. 2023. PMID: 36875581 Free PMC article. Review.
-
RiboNT: A Noise-Tolerant Predictor of Open Reading Frames from Ribosome-Protected Footprints.Life (Basel). 2021 Jul 16;11(7):701. doi: 10.3390/life11070701. Life (Basel). 2021. PMID: 34357073 Free PMC article.
-
D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery.Biology (Basel). 2024 Jul 26;13(8):563. doi: 10.3390/biology13080563. Biology (Basel). 2024. PMID: 39194501 Free PMC article.
-
Using AnABlast for intergenic sORF prediction in the Caenorhabditis elegans genome.Bioinformatics. 2020 Dec 8;36(19):4827-4832. doi: 10.1093/bioinformatics/btaa608. Bioinformatics. 2020. PMID: 32614398 Free PMC article.
-
The Emerging World of Small ORFs.Trends Plant Sci. 2016 Apr;21(4):317-328. doi: 10.1016/j.tplants.2015.11.005. Epub 2015 Dec 10. Trends Plant Sci. 2016. PMID: 26684391 Review.
Cited by
-
The Emerging Role of uORF-Encoded uPeptides and HLA uLigands in Cellular and Tumor Biology.Cancers (Basel). 2022 Dec 7;14(24):6031. doi: 10.3390/cancers14246031. Cancers (Basel). 2022. PMID: 36551517 Free PMC article. Review.
-
Improved super-resolution ribosome profiling reveals prevalent translation of upstream ORFs and small ORFs in Arabidopsis.Plant Cell. 2024 Feb 26;36(3):510-539. doi: 10.1093/plcell/koad290. Plant Cell. 2024. PMID: 38000896 Free PMC article.
-
slORFfinder: a tool to detect open reading frames resulting from trans-splicing of spliced leader sequences.Brief Bioinform. 2023 Jan 19;24(1):bbac610. doi: 10.1093/bib/bbac610. Brief Bioinform. 2023. PMID: 36611257 Free PMC article.
-
What, where, and how: Regulation of translation and the translational landscape in plants.Plant Cell. 2024 May 1;36(5):1540-1564. doi: 10.1093/plcell/koad197. Plant Cell. 2024. PMID: 37437121 Free PMC article. Review.
-
Identification of short open reading frames in plant genomes.Front Plant Sci. 2023 Feb 15;14:1094715. doi: 10.3389/fpls.2023.1094715. eCollection 2023. Front Plant Sci. 2023. PMID: 36875581 Free PMC article. Review.
References
-
- Calviello L, Mukherjee N, Wyler E, et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 2016;13:165–70. - PubMed
-
- Calviello L, Ohler U. Beyond read-counts: ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet 2017;33:728–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

