Absolute quantification of plasma mitochondrial DNA by droplet digital PCR marks COVID-19 severity over time during intensive care unit admissions
- PMID: 35699291
- PMCID: PMC9273271
- DOI: 10.1152/ajplung.00128.2022
Absolute quantification of plasma mitochondrial DNA by droplet digital PCR marks COVID-19 severity over time during intensive care unit admissions
Abstract
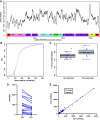
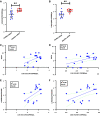
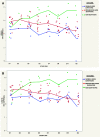
Increased plasma mitochondrial DNA concentrations are associated with poor outcomes in multiple critical illnesses, including COVID-19. However, current methods of cell-free mitochondrial DNA quantification in plasma are time-consuming and lack reproducibility. Here, we used next-generation sequencing to characterize the size and genome location of circulating mitochondrial DNA in critically ill subjects with COVID-19 to develop a facile and optimal method of quantification by droplet digital PCR. Sequencing revealed a large percentage of small mitochondrial DNA fragments in plasma with wide variability in coverage by genome location. We identified probes for the mitochondrial DNA genes, cytochrome B and NADH dehydrogenase 1, in regions of relatively high coverage that target small sequences potentially missed by other methods. Serial assessments of absolute mitochondrial DNA concentrations were then determined in plasma from 20 critically ill subjects with COVID-19 without a DNA isolation step. Mitochondrial DNA concentrations on the day of enrollment were increased significantly in patients with moderate or severe acute respiratory distress syndrome (ARDS) compared with those with no or mild ARDS. Comparisons of mitochondrial DNA concentrations over time between patients with no/mild ARDS who survived, patients with moderate/severe ARDS who survived, and nonsurvivors showed the highest concentrations in patients with more severe disease. Absolute mitochondrial DNA quantification by droplet digital PCR is time-efficient and reproducible; thus, we provide a valuable tool and rationale for future studies evaluating mitochondrial DNA as a real-time biomarker to guide clinical decision-making in critically ill subjects with COVID-19.
Keywords: ARDS; COVID-19; mitochondrial DNA.
Conflict of interest statement
G. Cutter has the following conflicts of interest to disclose: Data and Safety Monitoring Boards: AI Therapeutics, AMO Pharma, Astra-Zeneca, Avexis Pharmaceuticals, Biolinerx, Brainstorm Cell Therapeutics, Bristol Meyers Squibb/Celgene, CSL Behring, Galmed Pharmaceuticals, Green Valley Pharma, Horizon Pharmaceuticals, Immunic, Mapi Pharmaceuticals LTD, Merck, Mitsubishi Tanabe Pharma Holdings, Opko Biologics, Prothena Biosciences, Novartis, Regeneron, Sanofi-Aventis, Reata Pharmaceuticals, NHLBI (Protocol Review Committee), University of Texas Southwestern, University of Pennsylvania, Visioneering Technologies, Inc. Consulting or Advisory Boards: Alexion, Antisense Therapeutics, Biogen, Clinical Trial Solutions LLC, Genzyme, Genentech, GW Pharmaceuticals, Immunic, Klein-Buendel Incorporated, Merck/Serono, Osmotica Pharmaceuticals, Perception Neurosciences, Protalix Biotherapeutics, Recursion/Cerexis Pharmaceuticals, Regeneron, Roche, SAB Biotherapeutics. G. Cutter is employed by the University of Alabama at Birmingham and is President of Pythagoras, Inc., a private consulting company located in Birmingham, Alabama. J. H. Ix leads an Investigator Initiated Research Grant supported by Baxter International, has served on Advisory Boards for Akebia, AstraZeneca, and Bayer, and serves as a member of a Data and Safety Monitoring Board for Sanifit International. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
Figures
References
-
- Guérin C, Reignier J, Richard JC, Beuret P, Gacouin A, Boulain T, Mercier E, Badet M, Mercat A, Baudin O, Clavel M, Chatellier D, Jaber S, Rosselli S, Mancebo J, Sirodot M, Hilbert G, Bengler C, Richecoeur J, Gainnier M, Bayle F, Bourdin G, Leray V, Girard R, Baboi L, Ayzac L. PROSEVA Study Group. Prone positioning in severe acute respiratory distress syndrome. N Engl J Med 368: 2159–2168, 2013. doi: 10.1056/NEJMoa1214103. - DOI - PubMed
-
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520: 553–557, 2015. doi: 10.1038/nature14156. - DOI - PMC - PubMed
-
- Krychtiuk KA, Ruhittel S, Hohensinner PJ, Koller L, Kaun C, Lenz M, Bauer B, Wutzlhofer L, Draxler DF, Maurer G, Huber K, Wojta J, Heinz G, Niessner A, Speidl WS. Mitochondrial DNA and toll-like receptor-9 are associated with mortality in critically ill patients. Crit Care Med 43: 2633–2641, 2015. doi: 10.1097/CCM.0000000000001311. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM127823/GM/NIGMS NIH HHS/United States
- T32 HL134632/HL/NHLBI NIH HHS/United States
- UL1 TR001442/TR/NCATS NIH HHS/United States
- R01 HL113614/HL/NHLBI NIH HHS/United States
- T32 CA009370/CA/NCI NIH HHS/United States
- K08 NS109200/NS/NINDS NIH HHS/United States
- IK2 BX004338/BX/BLRD VA/United States
- P30 DK079337/DK/NIDDK NIH HHS/United States
- F31 AG062099/AG/NIA NIH HHS/United States
- K24 DK110427/DK/NIDDK NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- R01 AR069876/AR/NIAMS NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

