Predicting Protein-Ligand Docking Structure with Graph Neural Network
- PMID: 35699430
- PMCID: PMC10279412
- DOI: 10.1021/acs.jcim.2c00127
Predicting Protein-Ligand Docking Structure with Graph Neural Network
Abstract
Modern day drug discovery is extremely expensive and time consuming. Although computational approaches help accelerate and decrease the cost of drug discovery, existing computational software packages for docking-based drug discovery suffer from both low accuracy and high latency. A few recent machine learning-based approaches have been proposed for virtual screening by improving the ability to evaluate protein-ligand binding affinity, but such methods rely heavily on conventional docking software to sample docking poses, which results in excessive execution latencies. Here, we propose and evaluate a novel graph neural network (GNN)-based framework, MedusaGraph, which includes both pose-prediction (sampling) and pose-selection (scoring) models. Unlike the previous machine learning-centric studies, MedusaGraph generates the docking poses directly and achieves from 10 to 100 times speedup compared to state-of-the-art approaches, while having a slightly better docking accuracy.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
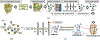

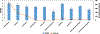

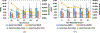
References
-
- Ruddigkeit L; Van Deursen R; Blum LC; Reymond J-L Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J. Chem. Inf. Model 2012, 52, 2864–2875. - PubMed
-
- Declerck PJ; Darendeliler F; Goth M; Kolouskova S; Micle I; Noordam C; Peterkova V; Volevodz NN; Zapletalová J; Ranke MB Biosimilars: controversies as illustrated by rhGH. Curr. Med. Res. Opin 2010, 26, 1219–1229. - PubMed
-
- Di Stasi A; Tey S-K; Dotti G; Fujita Y; Kennedy-Nasser A; Martinez C; Straathof K; Liu E; Durett AG; Grilley B; Liu H; Cruz CR; Savoldo B; Gee AP; Schindler J; Krance RA; Heslop HE; Spencer DM; Rooney CM; Brenner MK Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med 2011, 365, 1673–1683. - PMC - PubMed
-
- Convertino M; Das J; Dokholyan NV Pharmacological chaperones: design and development of new therapeutic strategies for the treatment of conformational diseases. ACS Chem. Biol 2016, 11, 1471–1489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

