Immune boosting by B.1.1.529 ( Omicron) depends on previous SARS-CoV-2 exposure
- PMID: 35699621
- PMCID: PMC9210451
- DOI: 10.1126/science.abq1841
Immune boosting by B.1.1.529 ( Omicron) depends on previous SARS-CoV-2 exposure
Abstract
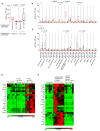
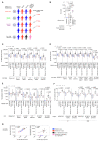
The Omicron, or Pango lineage B.1.1.529, variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) carries multiple spike mutations with high transmissibility and partial neutralizing antibody (nAb) escape. Vaccinated individuals show protection against severe disease, often attributed to primed cellular immunity. We investigated T and B cell immunity against B.1.1.529 in triple BioNTech BNT162b2 messenger RNA-vaccinated health care workers (HCWs) with different SARS-CoV-2 infection histories. B and T cell immunity against previous variants of concern was enhanced in triple-vaccinated individuals, but the magnitude of T and B cell responses against B.1.1.529 spike protein was reduced. Immune imprinting by infection with the earlier B.1.1.7 (Alpha) variant resulted in less durable binding antibody against B.1.1.529. Previously infection-naïve HCWs who became infected during the B.1.1.529 wave showed enhanced immunity against earlier variants but reduced nAb potency and T cell responses against B.1.1.529 itself. Previous Wuhan Hu-1 infection abrogated T cell recognition and any enhanced cross-reactive neutralizing immunity on infection with B.1.1.529.
Figures




References
-
- Elliott P., Bodinier B., Eales O., Wang H., Haw D., Elliott J., Whitaker M., Jonnerby J., Tang D., Walters C. E., Atchison C., Diggle P. J., Page A. J., Trotter A. J., Ashby D., Barclay W., Taylor G., Ward H., Darzi A., Cooke G. S., Chadeau-Hyam M., Donnelly C. A., Rapid increase in Omicron infections in England during December 2021: REACT-1 study. Science 375, 1406–1411 (2022). 10.1126/science.abn8347 - DOI - PMC - PubMed
-
- Madhi S. A., Kwatra G., Myers J. E., Jassat W., Dhar N., Mukendi C. K., Nana A. J., Blumberg L., Welch R., Ngorima-Mabhena N., Mutevedzi P. C., Population immunity and Covid-19 severity with omicron variant in South Africa. N. Engl. J. Med. 386, 1314–1326 (2022). 10.1056/NEJMoa2119658 - DOI - PMC - PubMed
-
- Hui K. P. Y., Ho J. C. W., Cheung M. C., Ng K. C., Ching R. H. H., Lai K. L., Kam T. T., Gu H., Sit K. Y., Hsin M. K. Y., Au T. W. K., Poon L. L. M., Peiris M., Nicholls J. M., Chan M. C. W., SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603, 715–720 (2022). 10.1038/s41586-022-04479-6 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

