Susceptibility of SARS-CoV-2 Omicron Variants to Therapeutic Monoclonal Antibodies: Systematic Review and Meta-analysis
- PMID: 35700134
- PMCID: PMC9430471
- DOI: 10.1128/spectrum.00926-22
Susceptibility of SARS-CoV-2 Omicron Variants to Therapeutic Monoclonal Antibodies: Systematic Review and Meta-analysis
Abstract
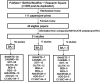
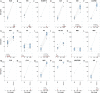
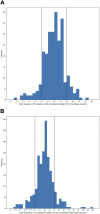
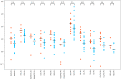
SARS-CoV-2 Omicron variants contain many mutations in its spike receptor-binding domain, the target of all authorized monoclonal antibodies (MAbs). Determining the extent to which Omicron variants reduced MAb susceptibility is critical to preventing and treating COVID-19. We systematically reviewed PubMed and three preprint servers, last updated 11 April 2022, for the in vitro activity of authorized MAbs against the Omicron variants. Fifty-one studies were eligible, including 50 containing Omicron BA.1 susceptibility data and 17 containing Omicron BA.2 susceptibility data. The first two authorized MAb combinations, bamlanivimab/etesevimab and casirivimab/imdevimab, were largely inactive against the Omicron BA.1 and BA.2 variants. In 34 studies, sotrovimab displayed a median 4.0-fold (interquartile range [IQR]: 2.6 to 6.9) reduction in activity against Omicron BA.1, and in 12 studies, it displayed a median 17-fold (IQR: 13 to 30) reduction in activity against Omicron BA.2. In 15 studies, the combination cilgavimab/tixagevimab displayed a median 86-fold (IQR: 27 to 151) reduction in activity against Omicron BA.1, and in six studies, it displayed a median 5.4-fold (IQR: 3.7 to 6.9) reduction in activity against Omicron BA.2. In eight studies against Omicron BA.1 and six studies against Omicron BA.2, bebtelovimab displayed no reduction in activity. Disparate results between assays were common. For authorized MAbs, 51/268 (19.0%) results for wild-type control variants and 78/348 (22.4%) results for Omicron BA.1 and BA.2 variants were more than 4-fold below or 4-fold above the median result for that MAb. Highly disparate results between published assays indicate a need for improved MAb susceptibility test standardization or interassay calibration. IMPORTANCE Monoclonal antibodies (MAbs) targeting the SARS-CoV-2 spike protein are among the most effective measures for preventing and treating COVID-19. However, SARS-CoV-2 Omicron variants contain many mutations in their spike receptor-binding domains, the target of all authorized MAbs. Therefore, determining the extent to which Omicron variants reduced MAb susceptibility is critical to preventing and treating COVID-19. We identified 51 studies that reported the in vitro susceptibility of the two main Omicron variants BA.1 and BA.2 to therapeutic MAbs in advanced clinical development, including eight authorized individual MAbs and three authorized MAb combinations. We estimated the degree to which different MAbs displayed reduced activity against Omicron variants. The marked loss of activity of many MAbs against Omicron variants underscores the importance of developing MAbs that target conserved regions of spike. Highly disparate results between assays indicate the need for improved MAb susceptibility test standardization.
Keywords: COVID-19; Omicron variant; SARS-CoV-2; antiviral therapy; monoclonal antibody; multidrug resistance; neutralization; spike protein.
Conflict of interest statement
The authors declare a conflict of interest. RWS served on a Vir Biotechnologies/GlaxoSmithKline advisory board in 2021. However, this did not influence the conduct of the submitted study.
Figures


Similar articles
-
Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies.Nat Med. 2022 Jun;28(6):1297-1302. doi: 10.1038/s41591-022-01792-5. Epub 2022 Mar 23. Nat Med. 2022. PMID: 35322239
-
Clinical efficacy and in vitro neutralization capacity of monoclonal antibodies for severe acute respiratory syndrome coronavirus 2 delta and omicron variants.J Med Virol. 2022 Oct;94(10):5038-5043. doi: 10.1002/jmv.27916. Epub 2022 Jun 11. J Med Virol. 2022. PMID: 35662058 Free PMC article.
-
Molecular Characterization of AZD7442 (Tixagevimab-Cilgavimab) Neutralization of SARS-CoV-2 Omicron Subvariants.Microbiol Spectr. 2023 Mar 6;11(2):e0033323. doi: 10.1128/spectrum.00333-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 36877050 Free PMC article.
-
Impact of SARS-CoV-2 variant mutations on susceptibility to monoclonal antibodies and antiviral drugs: a non-systematic review, April 2022 to October 2024.Euro Surveill. 2025 Mar;30(10):2400252. doi: 10.2807/1560-7917.ES.2025.30.10.2400252. Euro Surveill. 2025. PMID: 40084420 Free PMC article. Review.
-
Immune treatment in COVID-19.Rev Esp Quimioter. 2022 Apr;35 Suppl 1(Suppl 1):59-63. doi: 10.37201/req/s01.14.2022. Epub 2022 Apr 22. Rev Esp Quimioter. 2022. PMID: 35488829 Free PMC article. Review.
Cited by
-
Comparative Effectiveness of Antivirals and Monoclonal Antibodies for Treating COVID-19 Patients Infected With Omicron Variant: A Systematic Review and Network Meta-Analysis.Influenza Other Respir Viruses. 2024 Dec;18(12):e70065. doi: 10.1111/irv.70065. Influenza Other Respir Viruses. 2024. PMID: 39722466 Free PMC article.
-
A comprehensive insight into current control of COVID-19: Immunogenicity, vaccination, and treatment.Biomed Pharmacother. 2022 Sep;153:113499. doi: 10.1016/j.biopha.2022.113499. Epub 2022 Aug 2. Biomed Pharmacother. 2022. PMID: 36076589 Free PMC article. Review.
-
Qualitative monitoring of SARS-CoV-2 mRNA vaccination in humans using droplet microfluidics.JCI Insight. 2023 Jul 10;8(13):e166602. doi: 10.1172/jci.insight.166602. JCI Insight. 2023. PMID: 37252802 Free PMC article.
-
Prevention and treatment strategies for kidney transplant recipients in the context of long-term existence of COVID-19.Front Med (Lausanne). 2024 Apr 3;11:1287836. doi: 10.3389/fmed.2024.1287836. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38633308 Free PMC article. Review.
-
Safety and efficacy of tixagevimab/cilgavimab for pre-exposure prophylaxis in kidney transplant recipients: a multicenter retrospective cohort study.J Nephrol. 2024 Jul;37(6):1539-1550. doi: 10.1007/s40620-024-01889-9. Epub 2024 May 23. J Nephrol. 2024. PMID: 38780697 Free PMC article.
References
-
- Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP, Bjorkman PJ. 2020. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588:682–687. doi:10.1038/s41586-020-2852-1. - DOI - PMC - PubMed
-
- Dejnirattisai W, Zhou D, Ginn HM, Duyvesteyn HME, Supasa P, Case JB, Zhao Y, Walter TS, Mentzer AJ, Liu C, Wang B, Paesen GC, Slon-Campos J, López-Camacho C, Kafai NM, Bailey AL, Chen RE, Ying B, Thompson C, Bolton J, Fyfe A, Gupta S, Tan TK, Gilbert-Jaramillo J, James W, Knight M, Carroll MW, Skelly D, Dold C, Peng Y, Levin R, Dong T, Pollard AJ, Knight JC, Klenerman P, Temperton N, Hall DR, Williams MA, Paterson NG, Bertram FKR, Siebert CA, Clare DK, Howe A, Radecke J, Song Y, Townsend AR, Huang K-YA, Fry EE, Mongkolsapaya J, Diamond MS, Ren J, Stuart DI, Screaton GR. 2021. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184:2183–2200.e22. doi:10.1016/j.cell.2021.02.032. - DOI - PMC - PubMed
-
- COVID-19 Treatment Guidelines Panel. 2022. Coronavirus disease 2019 (COVID-19) treatment guidelines. National Institutes of Health, Bethesda, MD. https://www.covid19treatmentguidelines.nih.gov/. Accessed 7 January 2022.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

