Gardnerella vaginalis clades in pregnancy: New insights into the interactions with the vaginal microbiome
- PMID: 35700195
- PMCID: PMC9197028
- DOI: 10.1371/journal.pone.0269590
Gardnerella vaginalis clades in pregnancy: New insights into the interactions with the vaginal microbiome
Abstract
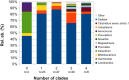
Gardnerella vaginalis (GV) is an anaerobic bacterial species involved in the pathogenesis of bacterial vaginosis (BV), a condition of vaginal dysbiosis associated with adverse pregnancy outcomes. GV strains are categorized into four clades, characterized by a different ability to produce virulence factors, such as sialidase. We investigated the distribution of GV clades and sialidase genes in the vaginal ecosystem of a cohort of pregnant women, assessing the correlations between GV clades and the whole vaginal microbiome. A total of 61 Caucasian pregnant women were enrolled. Their vaginal swabs, collected both at the first and third trimester of pregnancy, were used for (i) evaluation of the vaginal status by Nugent score, (ii) vaginal microbiome profiling by 16S rRNA sequencing, (iii) detection and quantification of GV clades and sialidase A gene by qPCR assays. DNA of at least one GV clade was detected in most vaginal swabs, with clade 4 being the most common one. GV clade 2, together with the presence of multiple clades (>2 simultaneously), were significantly associated with a BV condition. Significantly higher GV loads and sialidase gene levels were found in BV cases, compared to the healthy status. Clade 2 was related to the major shifts in the vaginal microbial composition, with a decrease in Lactobacillus and an increase in several BV-related taxa. As the number of GV clades detected simultaneously increased, a group of BV-associated bacteria tended to increase as well, while Bifidobacterium tended to decrease. A negative correlation between sialidase gene levels and Lactobacillus, and a positive correlation with Gardnerella, Atopobium, Prevotella, Megasphaera, and Sneathia were observed. Our results added knowledge about the interactions of GV clades with the inhabitants of the vaginal microbiome, possibly helping to predict the severity of BV and opening new perspectives for the prevention of pregnancy-related complications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

