Influenza A virus undergoes compartmentalized replication in vivo dominated by stochastic bottlenecks
- PMID: 35701424
- PMCID: PMC9197827
- DOI: 10.1038/s41467-022-31147-0
Influenza A virus undergoes compartmentalized replication in vivo dominated by stochastic bottlenecks
Abstract
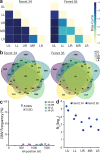
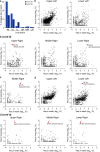
Transmission of influenza A viruses (IAV) between hosts is subject to numerous physical and biological barriers that impose genetic bottlenecks, constraining viral diversity and adaptation. The bottlenecks within hosts and their potential impacts on evolutionary pathways taken during infection are poorly understood. To address this, we created highly diverse IAV libraries bearing molecular barcodes on two gene segments, enabling high-resolution tracking and quantification of unique virus lineages within hosts. Here we show that IAV infection in lungs is characterized by multiple within-host bottlenecks that result in "islands" of infection in lung lobes, each with genetically distinct populations. We perform site-specific inoculation of barcoded IAV in the upper respiratory tract of ferrets and track viral diversity as infection spreads to the trachea and lungs. We detect extensive compartmentalization of discrete populations within lung lobes. Bottleneck events and localized replication stochastically sample individual viruses from the upper respiratory tract or the trachea that become the dominant genotype in a particular lobe. These populations are shaped strongly by founder effects, with limited evidence for positive selection. The segregated sites of replication highlight the jackpot-style events that contribute to within-host influenza virus evolution and may account for low rates of intrahost adaptation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

