Brain-wide reconstruction of inhibitory circuits after traumatic brain injury
- PMID: 35701434
- PMCID: PMC9197933
- DOI: 10.1038/s41467-022-31072-2
Brain-wide reconstruction of inhibitory circuits after traumatic brain injury
Abstract
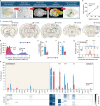
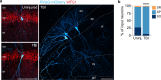
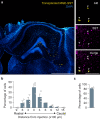
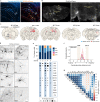
Despite the fundamental importance of understanding the brain's wiring diagram, our knowledge of how neuronal connectivity is rewired by traumatic brain injury remains remarkably incomplete. Here we use cellular resolution whole-brain imaging to generate brain-wide maps of the input to inhibitory neurons in a mouse model of traumatic brain injury. We find that somatostatin interneurons are converted into hyperconnected hubs in multiple brain regions, with rich local network connections but diminished long-range inputs, even at areas not directly damaged. The loss of long-range input does not correlate with cell loss in distant brain regions. Interneurons transplanted into the injury site receive orthotopic local and long-range input, suggesting the machinery for establishing distant connections remains intact even after a severe injury. Our results uncover a potential strategy to sustain and optimize inhibition after traumatic brain injury that involves spatial reorganization of the direct inputs to inhibitory neurons across the brain.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
iDISCO Inferno: Mapping Connectivity Changes Following TBI.Epilepsy Curr. 2023 Jan 18;23(2):127-129. doi: 10.1177/15357597221149263. eCollection 2023 Mar-Apr. Epilepsy Curr. 2023. PMID: 37122404 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

