Identification of a novel immune gene panel in tongue squamous cell carcinoma
- PMID: 35702068
- PMCID: PMC9185061
Identification of a novel immune gene panel in tongue squamous cell carcinoma
Abstract
Background: Tongue squamous cell carcinoma (TSCC) is one of the most common oral cancers. Immune activity is significantly related to the initiation and progression of TSCC. Systemic analysis of the immunogenomic landscape and identification of crucial immune-related genes (IRGs) would help understanding of TSCC. Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) provide multiple TSCC cases for use in an integrated immunogenomic study.
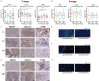
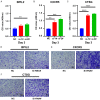
Methods: Immune landscape of TSCC was depicted by expression microarray data from GSE13601 and GSE34105. Univariate Cox analysis, in combination with survival analysis, was applied to select candidate IRGs with significant survival value. Survival predicting models were constructed by multivariate Cox regression and logistic regression analysis. Unsupervised clustering analysis was used to construct an immune gene panel based on prognostic IRGs to distinguish TSCC subgroups with different prognostic outcomes. Finally, IHC staining was performed to validate the clinical value of this immune-gene panel.
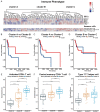
Results: Differentially expressed IRGs were identified in two TSCC microarray datasets. Functional enrichment analysis revealed that ontology terms associated with variations in T cell function, were highly enriched. Infiltration status of activated CD8+ T cells, central memory CD4+ T cells and type 17 T helper cells, had great prognostic value for TSCC progression. Unsupervised clustering analysis was further performed to classify TSCC patients into three subgroups. CTSG, CXCL13, and VEGFA were finally combined together to form an immune-gene panel, todistinguish different TSCC subgroups. IHC staining of TSCC sections further validated the clinical efficiency of the immune-gene panel consisting of prognostic IRGs to distinguish TSCC patients.
Conclusion: VEGFA, CXCL13, and CTSG, correlated with T cell infiltration and prognostic outcome. They were screened to form an immune-gene panel to identify TSCC subgroups with different prognostic outcomes. Clinical IHC further validated the efficacy of this immune-gene panel to evaluate aggressiveness of TSCC development.
Keywords: CTSG; CXCL13; Oral squamous cell carcinoma; VEGFA; immune-related genes; tongue squamous cell carcinoma.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures









Similar articles
-
Identification and Validation of an Invasion-Related Disease-Free Survival Prognostic Model for Tongue Squamous Cell Carcinoma.Oncology. 2025;103(3):237-252. doi: 10.1159/000540977. Epub 2024 Sep 20. Oncology. 2025. PMID: 39307124
-
Development and validation of a predictive model for immune-related genes in patients with tongue squamous cell carcinoma.Open Life Sci. 2022 Dec 15;17(1):1657-1668. doi: 10.1515/biol-2022-0469. eCollection 2022. Open Life Sci. 2022. PMID: 36567723 Free PMC article.
-
Novel Prognostic Model Construction of Tongue Squamous Cell Carcinoma Based on Apigenin-Associated Genes.Front Biosci (Landmark Ed). 2024 Feb 6;29(2):65. doi: 10.31083/j.fbl2902065. Front Biosci (Landmark Ed). 2024. PMID: 38420803
-
Development of an Immunogenomic Landscape-Based Prognostic Index of Head and Neck Squamous Cell Carcinoma.Front Mol Biosci. 2020 Nov 24;7:586344. doi: 10.3389/fmolb.2020.586344. eCollection 2020. Front Mol Biosci. 2020. PMID: 33330624 Free PMC article.
-
A narrative review on machine learning in diagnosis and prognosis prediction for tongue squamous cell carcinoma.Transl Cancer Res. 2022 Dec;11(12):4409-4415. doi: 10.21037/tcr-22-1669. Transl Cancer Res. 2022. PMID: 36644177 Free PMC article. Review.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Mannelli G, Arcuri F, Agostini T, Innocenti M, Raffaini M, Spinelli G. Classification of tongue cancer resection and treatment algorithm. J Surg Oncol. 2018;117:1092–1099. - PubMed
-
- Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26:645–662. - PubMed
-
- Gupta B, Johnson NW, Kumar N. Global epidemiology of head and neck cancers: a continuing challenge. Oncology. 2016;91:13–23. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
