The transient expression of recombinant proteins in plant cell packs facilitates stable isotope labelling for NMR spectroscopy
- PMID: 35702941
- PMCID: PMC9491462
- DOI: 10.1111/pbi.13873
The transient expression of recombinant proteins in plant cell packs facilitates stable isotope labelling for NMR spectroscopy
Abstract
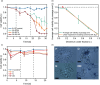
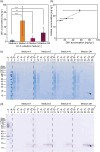
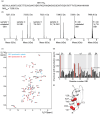
Nuclear magnetic resonance (NMR) spectroscopy can be used to determine the structure, dynamics and interactions of proteins. However, protein NMR requires stable isotope labelling for signal detection. The cells used for the production of recombinant proteins must therefore be grown in medium containing isotopically labelled substrates. Stable isotope labelling is well established in Escherichia coli, but bacteria are only suitable for the production of simple proteins without post-translational modifications. More complex proteins require eukaryotic production hosts, but their growth can be impaired by labelled media, thus reducing product yields and increasing costs. To address this limitation, we used media supplemented with isotope-labelled substrates to cultivate the tobacco-derived cell line BY-2, which was then cast into plant cell packs (PCPs) for the transient expression of a labelled version of the model protein GB1. Mass spectrometry confirmed the feasibility of isotope labelling with 15 N and 2 H using this approach. The resulting NMR spectrum featured a signal dispersion comparable to recombinant GB1 produced in E. coli. PCPs therefore offer a rapid and cost-efficient alternative for the production of isotope-labelled proteins for NMR analysis, especially suitable for complex proteins that cannot be produced in microbial systems.
Keywords: alternative expression host; defined cultivation media; isotope labelling; plant cell culture; structural analysis.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures
References
-
- Arfi, Z.A. , Hellwig, S. , Drossard, J. , Fischer, R. and Buyel, J.F. (2016) Polyclonal antibodies for specific detection of tobacco host cell proteins can be efficiently generated following RuBisCO depletion and the removal of endotoxins. Biotechnol. J. 11(4), 507–518. - PubMed
-
- Bindschedler, L.V. , Palmblad, M. and Cramer, R. (2008) Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics; an oxidative stress case study. Phytochemistry, 69(10), 1962–1972. - PubMed
-
- Bouvignies, G. , Markwick, P. , Brüschweiler, R. and Blackledge, M. (2006) Simultaneous determination of protein backbone structure and dynamics from residual dipolar couplings. J. Am. Chem. Soc. 128(47), 15100–15101. - PubMed
-
- Buyel, J.F. , Kaever, T. , Buyel, J.J. and Fischer, R. (2013) Predictive models for the accumulation of a fluorescent marker protein in tobacco leaves according to the promoter/5′UTR combination. Biotechnol. Bioeng. 110(2), 471–482. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

