miR-218 affects the ECM composition and cell biomechanical properties of glioblastoma cells
- PMID: 35702951
- PMCID: PMC9279592
- DOI: 10.1111/jcmm.17428
miR-218 affects the ECM composition and cell biomechanical properties of glioblastoma cells
Abstract
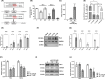
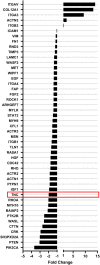
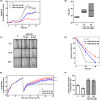
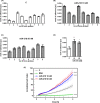
Glioblastoma (GBM) is the most common malignant brain tumour. GBM cells have the ability to infiltrate into the surrounding brain tissue, which results in a significant decrease in the patient's survival rate. Infiltration is a consequence of the low adhesion and high migration of the tumour cells, two features being associated with the highly remodelled extracellular matrix (ECM). In this study, we report that ECM composition is partially regulated at the post-transcriptional level by miRNA. Particularly, we show that miR-218, a well-known miRNA suppressor, is involved in the direct regulation of ECM components, tenascin-C (TN-C) and syndecan-2 (SDC-2). We demonstrated that the overexpression of miR-218 reduces the mRNA and protein expression levels of TN-C and SDC-2, and subsequently influences biomechanical properties of GBM cells. Atomic force microscopy (AFM) and real-time migration analysis revealed that miR-218 overexpression impairs the migration potential and enhances the adhesive properties of cells. AFM analysis followed by F-actin staining demonstrated that the expression level of miR-218 has an impact on cell stiffness and cytoskeletal reorganization. Global gene expression analysis showed deregulation of a number of genes involved in tumour cell motility and adhesion or ECM remodelling upon miR-218 treatment, suggesting further indirect interactions between the cells and ECM. The results demonstrated a direct impact of miR-218 reduction in GBM tumours on the qualitative ECM content, leading to changes in the rigidity of the ECM and GBM cells being conducive to increased invasiveness of GBM.
Keywords: AFM; ECM; GBM; glioblastoma; miR-218; tenascin-C.
© 2022 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures

Similar articles
-
Hypoxia-inducible miR-196a modulates glioblastoma cell proliferation and migration through complex regulation of NRAS.Cell Oncol (Dordr). 2021 Apr;44(2):433-451. doi: 10.1007/s13402-020-00580-y. Epub 2021 Jan 19. Cell Oncol (Dordr). 2021. PMID: 33469841
-
Expression of tenascin-C in various human brain tumors and its relevance for survival in patients with astrocytoma.Cancer. 2003 Dec 1;98(11):2430-9. doi: 10.1002/cncr.11796. Cancer. 2003. PMID: 14635078
-
miR-330-5p suppresses glioblastoma cell proliferation and invasiveness through targeting ITGA5.Biosci Rep. 2017 Jun 21;37(3):BSR20170019. doi: 10.1042/BSR20170019. Print 2017 Jun 30. Biosci Rep. 2017. Retraction in: Biosci Rep. 2021 Sep 30;41(9):BSR-2017-0019_RET. doi: 10.1042/BSR-2017-0019_RET. PMID: 28336765 Free PMC article. Retracted.
-
Biologically Active TNIIIA2 Region in Tenascin-C Molecule: A Major Contributor to Elicit Aggressive Malignant Phenotypes From Tumors/Tumor Stroma.Front Immunol. 2020 Dec 9;11:610096. doi: 10.3389/fimmu.2020.610096. eCollection 2020. Front Immunol. 2020. PMID: 33362799 Free PMC article. Review.
-
Integrative analysis of cell adhesion molecules in glioblastoma identified prostaglandin F2 receptor inhibitor (PTGFRN) as an essential gene.BMC Cancer. 2022 Jun 11;22(1):642. doi: 10.1186/s12885-022-09682-2. BMC Cancer. 2022. PMID: 35690717 Free PMC article. Review.
Cited by
-
C-Myc/H19/miR-29b axis downregulates nerve/glial (NG)2 expression in glioblastoma multiforme.Mol Ther Nucleic Acids. 2024 Jan 12;35(1):102120. doi: 10.1016/j.omtn.2024.102120. eCollection 2024 Mar 12. Mol Ther Nucleic Acids. 2024. PMID: 38318212 Free PMC article.
-
Mechanobiological insight into brain diseases based on mechanosensitive channels: Common mechanisms and clinical potential.CNS Neurosci Ther. 2024 Jun;30(6):e14809. doi: 10.1111/cns.14809. CNS Neurosci Ther. 2024. PMID: 38923822 Free PMC article. Review.
-
Discovery of miRNA-mRNA regulatory networks in glioblastoma reveals novel insights into tumor microenvironment remodeling.Sci Rep. 2024 Nov 11;14(1):27493. doi: 10.1038/s41598-024-78337-y. Sci Rep. 2024. PMID: 39528571 Free PMC article.
-
High Expression of miR-218-5p in the Peripheral Blood Stream and Tumor Tissues of Pediatric Patients with Sarcomas.Biochem Genet. 2025 Aug;63(4):3313-3328. doi: 10.1007/s10528-024-10873-8. Epub 2024 Jul 2. Biochem Genet. 2025. PMID: 38954213 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

