The C-terminal head domain of Burkholderia pseudomallei BpaC has a striking hydrophilic core with an extensive solvent network
- PMID: 35703459
- PMCID: PMC9543794
- DOI: 10.1111/mmi.14953
The C-terminal head domain of Burkholderia pseudomallei BpaC has a striking hydrophilic core with an extensive solvent network
Abstract
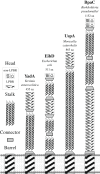
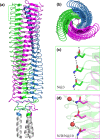
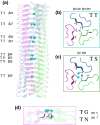
Gram-negative pathogens like Burkholderia pseudomallei use trimeric autotransporter adhesins such as BpaC as key molecules in their pathogenicity. Our 1.4 Å crystal structure of the membrane-proximal part of the BpaC head domain shows that the domain is exclusively made of left-handed parallel β-roll repeats. This, the largest such structure solved, has two unique features. First, the core, rather than being composed of the canonical hydrophobic Ile and Val, is made up primarily of the hydrophilic Thr and Asn, with two different solvent channels. Second, comparing BpaC to all other left-handed parallel β-roll structures showed that the position of the head domain in the protein correlates with the number and type of charged residues. In BpaC, only negatively charged residues face the solvent-in stark contrast to the primarily positive surface charge of the left-handed parallel β-roll "type" protein, YadA. We propose extending the definitions of these head domains to include the BpaC-like head domain as a separate subtype, based on its unusual sequence, position, and charge. We speculate that the function of left-handed parallel β-roll structures may differ depending on their position in the structure.
Keywords: Burkholderia pseudomallei; Type V secretion systems; bacterial adhesin; bacterial outer membrane proteins; melioidosis; protein conformation; β-sheet.
© 2022 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures






Similar articles
-
Recent advances in the understanding of trimeric autotransporter adhesins.Med Microbiol Immunol. 2020 Jun;209(3):233-242. doi: 10.1007/s00430-019-00652-3. Epub 2019 Dec 21. Med Microbiol Immunol. 2020. PMID: 31865405 Free PMC article. Review.
-
Systematic mutagenesis of genes encoding predicted autotransported proteins of Burkholderia pseudomallei identifies factors mediating virulence in mice, net intracellular replication and a novel protein conferring serum resistance.PLoS One. 2015 Apr 1;10(4):e0121271. doi: 10.1371/journal.pone.0121271. eCollection 2015. PLoS One. 2015. PMID: 25830295 Free PMC article.
-
Structure of a Burkholderia pseudomallei trimeric autotransporter adhesin head.PLoS One. 2010 Sep 20;5(9):e12803. doi: 10.1371/journal.pone.0012803. PLoS One. 2010. PMID: 20862217 Free PMC article.
-
Identification of a predicted trimeric autotransporter adhesin required for biofilm formation of Burkholderia pseudomallei.PLoS One. 2013 Nov 5;8(11):e79461. doi: 10.1371/journal.pone.0079461. eCollection 2013. PLoS One. 2013. PMID: 24223950 Free PMC article.
-
Type III Secretion in the Melioidosis Pathogen Burkholderia pseudomallei.Front Cell Infect Microbiol. 2017 Jun 15;7:255. doi: 10.3389/fcimb.2017.00255. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28664152 Free PMC article. Review.
Cited by
-
Structural basis for immune cell binding of Fusobacterium nucleatum via the trimeric autotransporter adhesin CbpF.Proc Natl Acad Sci U S A. 2025 Apr 15;122(15):e2418155122. doi: 10.1073/pnas.2418155122. Epub 2025 Apr 8. Proc Natl Acad Sci U S A. 2025. PMID: 40198705 Free PMC article.
References
-
- Agnew, C. , Borodina, E. , Zaccai, N.R. , Conners, R. , Burton, N.M. , Vicary, J.A. et al. (2011) Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding. Proceedings of the National Academy of Sciences of the United States of America, 108(37), 15174–15178. 10.1073/pnas.1106341108 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

