ubiF is involved in acid stress tolerance and symbiotic competitiveness in Rhizobium favelukesii LPU83
- PMID: 35704174
- PMCID: PMC9433554
- DOI: 10.1007/s42770-022-00780-8
ubiF is involved in acid stress tolerance and symbiotic competitiveness in Rhizobium favelukesii LPU83
Abstract
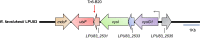
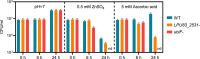
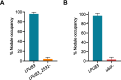
The acidity of soils significantly reduces the productivity of legumes mainly because of the detrimental effects of hydrogen ions on the legume plants, leading to the establishment of an inefficient symbiosis and poor biological nitrogen fixation. We recently reported the analysis of the fully sequenced genome of Rhizobium favelukesii LPU83, an alfalfa-nodulating rhizobium with a remarkable ability to grow, nodulate and compete in acidic conditions. To gain more insight into the genetic mechanisms leading to acid tolerance in R. favelukesii LPU83, we constructed a transposon mutant library and screened for mutants displaying a more acid-sensitive phenotype than the parental strain. We identified mutant Tn833 carrying a single-transposon insertion within LPU83_2531, an uncharacterized short ORF located immediately upstream from ubiF homolog. This gene encodes a protein with an enzymatic activity involved in the biosynthesis of ubiquinone. As the transposon was inserted near the 3' end of LPU83_2531 and these genes are cotranscribed as a part of the same operon, we hypothesized that the phenotype in Tn833 is most likely due to a polar effect on ubiF transcription.We found that a mutant in ubiF was impaired to grow at low pH and other abiotic stresses including 5 mM ascorbate and 0.500 mM Zn2+. Although the ubiF mutant retained the ability to nodulate alfalfa and Phaseolus vulgaris, it was unable to compete with the R. favelukesii LPU83 wild-type strain for nodulation in Medicago sativa and P. vulgaris, suggesting that ubiF is important for competitiveness. Here, we report for the first time an ubiF homolog being essential for nodulation competitiveness and tolerance to specific stresses in rhizobia.
Keywords: Acid tolerance; Competitiveness; Rhizobium favelukesii LPU83; Stress; ubiF.
© 2022. The Author(s) under exclusive licence to Sociedade Brasileira de Microbiologia.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Exopolysaccharide Characterization of Rhizobium favelukesii LPU83 and Its Role in the Symbiosis With Alfalfa.Front Plant Sci. 2021 Feb 10;12:642576. doi: 10.3389/fpls.2021.642576. eCollection 2021. Front Plant Sci. 2021. PMID: 33643369 Free PMC article.
-
The nodulation of alfalfa by the acid-tolerant Rhizobium sp. strain LPU83 does not require sulfated forms of lipochitooligosaccharide nodulation signals.J Bacteriol. 2011 Jan;193(1):30-9. doi: 10.1128/JB.01009-10. Epub 2010 Oct 22. J Bacteriol. 2011. PMID: 20971905 Free PMC article.
-
Proteomic Analysis of Rhizobium favelukesii LPU83 in Response to Acid Stress.J Proteome Res. 2019 Oct 4;18(10):3615-3629. doi: 10.1021/acs.jproteome.9b00275. Epub 2019 Sep 5. J Proteome Res. 2019. PMID: 31432679
-
Genetics of competition for nodulation of legumes.Annu Rev Microbiol. 1992;46:399-428. doi: 10.1146/annurev.mi.46.100192.002151. Annu Rev Microbiol. 1992. PMID: 1444262 Review.
-
[Nodulation competitiveness of nodule bacteria: Genetic control and adaptive significance].Prikl Biokhim Mikrobiol. 2017 Mar-Apr;53(2):127-35. Prikl Biokhim Mikrobiol. 2017. PMID: 29508968 Review. Russian.
Cited by
-
Bacterial persisters: molecular mechanisms and therapeutic development.Signal Transduct Target Ther. 2024 Jul 17;9(1):174. doi: 10.1038/s41392-024-01866-5. Signal Transduct Target Ther. 2024. PMID: 39013893 Free PMC article. Review.
-
Evaluation of Legume-Rhizobial Symbiotic Interactions Beyond Nitrogen Fixation That Help the Host Survival and Diversification in Hostile Environments.Microorganisms. 2023 May 31;11(6):1454. doi: 10.3390/microorganisms11061454. Microorganisms. 2023. PMID: 37374957 Free PMC article. Review.
References
-
- Cloutier J, Serge L, Yves C, Hani A (1996) Characterization and mutational analysis of nodHPQ genes of Rhizobium sp. strain N33. Mol Plant Microbe Interact Vol 9, 720–728. 10.1094/mpmi-9-0720, https://pubmed.ncbi.nlm.nih.gov/8870271/ - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

