Depletion of oocyte dynamin-related protein 1 shows maternal-effect abnormalities in embryonic development
- PMID: 35704569
- PMCID: PMC9200162
- DOI: 10.1126/sciadv.abl8070
Depletion of oocyte dynamin-related protein 1 shows maternal-effect abnormalities in embryonic development
Abstract
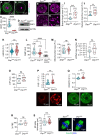
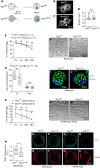
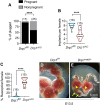
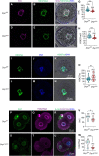
Eggs contain about 200,000 mitochondria that generate adenosine triphosphate and metabolites essential for oocyte development. Mitochondria also integrate metabolism and transcription via metabolites that regulate epigenetic modifiers, but there is no direct evidence linking oocyte mitochondrial function to the maternal epigenome and subsequent embryo development. Here, we have disrupted oocyte mitochondrial function via deletion of the mitochondrial fission factor Drp1. Fission-deficient oocytes exhibit a high frequency of failure in peri- and postimplantation development. This is associated with altered mitochondrial function, changes in the oocyte transcriptome and proteome, altered subcortical maternal complex, and a decrease in oocyte DNA methylation and H3K27me3. Transplanting pronuclei of fertilized Drp1 knockout oocytes to normal ooplasm fails to rescue embryonic lethality. We conclude that mitochondrial function plays a role in establishing the maternal epigenome, with serious consequences for embryo development.
Figures


References
-
- Kraus F., Ryan M. T., The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 130, 2953–2960 (2017). - PubMed
-
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y. I., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S., Mihara K., Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

