Construction of long non-coding RNA- and microRNA-mediated competing endogenous RNA networks in alcohol-related esophageal cancer
- PMID: 35704638
- PMCID: PMC9200351
- DOI: 10.1371/journal.pone.0269742
Construction of long non-coding RNA- and microRNA-mediated competing endogenous RNA networks in alcohol-related esophageal cancer
Abstract
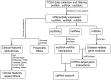
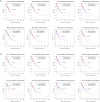
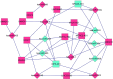
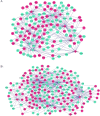
The current study aimed to explore the lncRNA-miRNA-mRNA networks associated with alcohol-related esophageal cancer (EC). RNA-sequencing and clinical data were downloaded from The Cancer Genome Atlas and the differentially expressed genes (DEGs), long non-coding RNAs (lncRNAs, DELs), and miRNAs (DEMs) in patients with alcohol-related and non-alcohol-related EC were identified. Prognostic RNAs were identified by performing Kaplan-Meier survival analyses. Weighted gene co-expression network analysis was employed to build the gene modules. The lncRNA-miRNA-mRNA competing endogenous RNA (ceRNA) networks were constructed based on our in silico analyses using data from miRcode, starBase, and miRTarBase databases. Functional enrichment analysis was performed for the genes in the identified ceRNA networks. A total of 906 DEGs, 40 DELs, and 52 DEMs were identified. There were eight lncRNAs and miRNAs each, including ST7-AS2 and miR-1269, which were significantly associated with the survival rate of patients with EC. Of the seven gene modules, the blue and turquoise modules were closely related to disease progression; the genes in this module were selected to construct the ceRNA networks. SNHG12-miR-1-ST6GAL1, SNHG3-miR-1-ST6GAL1, SPAG5-AS1-miR-133a-ST6GAL1, and SNHG12-hsa-miR-33a-ST6GA interactions, associated with the N-glycan biosynthesis pathway, may have key roles in alcohol-related EC. Thus, the identified biomarkers provide a novel insight into the molecular mechanism of alcohol-related EC.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: an integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks.Bioengineered. 2021 Dec;12(2):12821-12838. doi: 10.1080/21655979.2021.2003925. Bioengineered. 2021. PMID: 34898376 Free PMC article.
-
Predictions of the dysregulated competing endogenous RNA signature involved in the progression of human lung adenocarcinoma.Cancer Biomark. 2020;29(3):399-416. doi: 10.3233/CBM-200133. Cancer Biomark. 2020. PMID: 32741804
-
LINC01018 and SMIM25 sponged miR-182-5p in endometriosis revealed by the ceRNA network construction.Int J Immunopathol Pharmacol. 2020 Jan-Dec;34:2058738420976309. doi: 10.1177/2058738420976309. Int J Immunopathol Pharmacol. 2020. PMID: 33237828 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
LncRNA-miRNA-mRNA regulatory axes in endometrial cancer: a comprehensive overview.Arch Gynecol Obstet. 2022 Nov;306(5):1431-1447. doi: 10.1007/s00404-022-06423-5. Epub 2022 Feb 18. Arch Gynecol Obstet. 2022. PMID: 35182183 Review.
Cited by
-
The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions.Int J Mol Sci. 2022 Dec 13;23(24):15804. doi: 10.3390/ijms232415804. Int J Mol Sci. 2022. PMID: 36555445 Free PMC article. Review.
-
[Biological role of SPAG5 in the malignant proliferation of gastric cancer cells].Nan Fang Yi Ke Da Xue Xue Bao. 2024 Aug 20;44(8):1497-1507. doi: 10.12122/j.issn.1673-4254.2024.08.08. Nan Fang Yi Ke Da Xue Xue Bao. 2024. PMID: 39276045 Free PMC article. Chinese.
-
Ethanol- and PARP-Mediated Regulation of Ribosome-Associated Long Non-Coding RNA (lncRNA) in Pyramidal Neurons.Noncoding RNA. 2023 Nov 17;9(6):72. doi: 10.3390/ncrna9060072. Noncoding RNA. 2023. PMID: 37987368 Free PMC article.
-
Using ncRNAs as Tools in Cancer Diagnosis and Treatment-The Way towards Personalized Medicine to Improve Patients' Health.Int J Mol Sci. 2022 Aug 19;23(16):9353. doi: 10.3390/ijms23169353. Int J Mol Sci. 2022. PMID: 36012617 Free PMC article. Review.
-
Characterization of sialylation-related long noncoding RNAs to develop a novel signature for predicting prognosis, immune landscape, and chemotherapy response in colorectal cancer.Front Immunol. 2022 Oct 18;13:994874. doi: 10.3389/fimmu.2022.994874. eCollection 2022. Front Immunol. 2022. PMID: 36330513 Free PMC article.
References
-
- American Cancer S (2018) Cancer Facts and Figures 2018. American Cancer Society Atlanta
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

