Indoleamine 2,3-dioxygenase 1 activation in mature cDC1 promotes tolerogenic education of inflammatory cDC2 via metabolic communication
- PMID: 35704993
- PMCID: PMC9220322
- DOI: 10.1016/j.immuni.2022.05.013
Indoleamine 2,3-dioxygenase 1 activation in mature cDC1 promotes tolerogenic education of inflammatory cDC2 via metabolic communication
Abstract
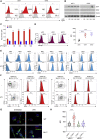
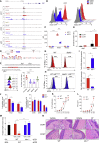
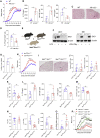
Conventional dendritic cells (cDCs), cDC1 and cDC2, act both to initiate immunity and maintain self-tolerance. The tryptophan metabolic enzyme indoleamine 2,3-dioxygenase 1 (IDO1) is used by cDCs in maintaining tolerance, but its role in different subsets remains unclear. At homeostasis, only mature CCR7+ cDC1 expressed IDO1 that was dependent on IRF8. Lipopolysaccharide treatment induced maturation and IDO1-dependent tolerogenic activity in isolated immature cDC1, but not isolated cDC2. However, both human and mouse cDC2 could induce IDO1 and acquire tolerogenic function when co-cultured with mature cDC1 through the action of cDC1-derived l-kynurenine. Accordingly, cDC1-specific inactivation of IDO1 in vivo exacerbated disease in experimental autoimmune encephalomyelitis. This study identifies a previously unrecognized metabolic communication in which IDO1-expressing cDC1 cells extend their immunoregulatory capacity to the cDC2 subset through their production of tryptophan metabolite l-kynurenine. This metabolic axis represents a potential therapeutic target in treating autoimmune demyelinating diseases.
Keywords: AhR; IDO1; IL-6; RelB; dendritic cells; immunotolerance; kynurenine; metabolites; neuroinflammation; tryptophan metabolism.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
cDC1 to cDC2: "Everything I do, Ido1 it for you".Immunity. 2022 Jun 14;55(6):967-970. doi: 10.1016/j.immuni.2022.05.015. Immunity. 2022. PMID: 35704997
References
-
- Ardouin L., Luche H., Chelbi R., Carpentier S., Shawket A., Montanana Sanchis F., Santa Maria C., Grenot P., Alexandre Y., Gregoire C., et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity. 2016;45:305–318. - PubMed
-
- Bacsi S.G., Hankinson O. Functional characterization of DNA-binding domains of the subunits of the heterodimeric aryl hydrocarbon receptor complex imputing novel and canonical basic helix-loop-helix protein-DNA interactions. J. Biol. Chem. 1996;271:8843–8850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

