Novel DNA Methylation Biomarker Panel for Detection of Esophageal Adenocarcinoma and High-Grade Dysplasia
- PMID: 35705525
- PMCID: PMC9444948
- DOI: 10.1158/1078-0432.CCR-22-0445
Novel DNA Methylation Biomarker Panel for Detection of Esophageal Adenocarcinoma and High-Grade Dysplasia
Abstract
Purpose: Current endoscopy-based screening and surveillance programs have not been proven effective at decreasing esophageal adenocarcinoma (EAC) mortality, creating an unmet need for effective molecular tests for early detection of this highly lethal cancer. We conducted a genome-wide methylation screen to identify novel methylation markers that distinguish EAC and high-grade dysplasia (HGD) from normal squamous epithelium (SQ) or nondysplastic Barrett's esophagus (NDBE).
Experimental design: DNA methylation profiling of samples from SQ, NDBE, HGD, and EAC was performed using HM450 methylation arrays (Illumina) and reduced-representation bisulfate sequencing. Ultrasensitive methylation-specific droplet digital PCR and next-generation sequencing (NGS)-based bisulfite-sequencing assays were developed to detect the methylation level of candidate CpGs in independent esophageal biopsy and endoscopic brushing samples.
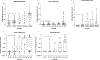
Results: Five candidate methylation markers were significantly hypermethylated in HGD/EAC samples compared with SQ or NDBE (P < 0.01) in both esophageal biopsy and endoscopic brushing samples. In an independent set of brushing samples used to construct biomarker panels, a four-marker panel (model 1) demonstrated sensitivity of 85.0% and 90.8% for HGD and EACs respectively, with 84.2% and 97.9% specificity for NDBE and SQ respectively. In a validation set of brushing samples, the panel achieved sensitivity of 80% and 82.5% for HGD and EAC respectively, at specificity of 67.6% and 96.3% for NDBE and SQ samples.
Conclusions: A novel DNA methylation marker panel differentiates HGD/EAC from SQ/NDBE. DNA-methylation-based molecular assays hold promise for the detection of HGD/EAC using esophageal brushing samples.
©2022 American Association for Cancer Research.
Figures


References
-
- Phoa KN, van Vilsteren FG, Weusten BL, et al. Radiofrequency ablation vs endoscopic surveillance for patients with Barrett esophagus and low-grade dysplasia: a randomized clinical trial. JAMA 2014;311:1209–17. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- K24 DK080941/DK/NIDDK NIH HHS/United States
- R35 CA197442/CA/NCI NIH HHS/United States
- P30 CA015704/CA/NCI NIH HHS/United States
- P30 DK097948/DK/NIDDK NIH HHS/United States
- U2C CA271902/CA/NCI NIH HHS/United States
- R01 CA220004/CA/NCI NIH HHS/United States
- R01 CA217992/CA/NCI NIH HHS/United States
- R01 CA247790/CA/NCI NIH HHS/United States
- P50 CA150964/CA/NCI NIH HHS/United States
- U01 CA086402/CA/NCI NIH HHS/United States
- R50 CA233042/CA/NCI NIH HHS/United States
- U01 CA182940/CA/NCI NIH HHS/United States
- U01 CA152756/CA/NCI NIH HHS/United States
- U54 CA163060/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

