Remote solid cancers rewire hepatic nitrogen metabolism via host nicotinamide-N-methyltransferase
- PMID: 35705545
- PMCID: PMC9200709
- DOI: 10.1038/s41467-022-30926-z
Remote solid cancers rewire hepatic nitrogen metabolism via host nicotinamide-N-methyltransferase
Abstract
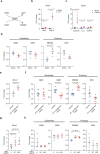
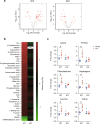
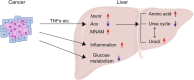
Cancers disrupt host homeostasis in various manners but the identity of host factors underlying such disruption remains largely unknown. Here we show that nicotinamide-N-methyltransferase (NNMT) is a host factor that mediates metabolic dysfunction in the livers of cancer-bearing mice. Multiple solid cancers distantly increase expression of Nnmt and its product 1-methylnicotinamide (MNAM) in the liver. Multi-omics analyses reveal suppression of the urea cycle accompanied by accumulation of amino acids, and enhancement of uracil biogenesis in the livers of cancer-bearing mice. Importantly, genetic deletion of Nnmt leads to alleviation of these metabolic abnormalities, and buffers cancer-dependent weight loss and reduction of the voluntary wheel-running activity. Our data also demonstrate that MNAM is capable of affecting urea cycle metabolites in the liver. These results suggest that cancers up-regulate the hepatic NNMT pathway to rewire liver metabolism towards uracil biogenesis rather than nitrogen disposal via the urea cycle, thereby disrupting host homeostasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

