Clinical genomic profiling in the management of patients with soft tissue and bone sarcoma
- PMID: 35705558
- PMCID: PMC9200814
- DOI: 10.1038/s41467-022-30496-0
Clinical genomic profiling in the management of patients with soft tissue and bone sarcoma
Abstract
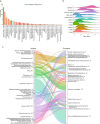
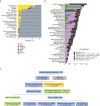
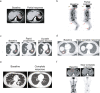
There are more than 70 distinct sarcomas, and this diversity complicates the development of precision-based therapeutics for these cancers. Prospective comprehensive genomic profiling could overcome this challenge by providing insight into sarcomas' molecular drivers. Through targeted panel sequencing of 7494 sarcomas representing 44 histologies, we identify highly recurrent and type-specific alterations that aid in diagnosis and treatment decisions. Sequencing could lead to refinement or reassignment of 10.5% of diagnoses. Nearly one-third of patients (31.7%) harbor potentially actionable alterations, including a significant proportion (2.6%) with kinase gene rearrangements; 3.9% have a tumor mutational burden ≥10 mut/Mb. We describe low frequencies of microsatellite instability (<0.3%) and a high degree of genome-wide loss of heterozygosity (15%) across sarcomas, which are not readily explained by homologous recombination deficiency (observed in 2.5% of cases). In a clinically annotated subset of 118 patients, we validate actionable genetic events as therapeutic targets. Collectively, our findings reveal the genetic landscape of human sarcomas, which may inform future development of therapeutics and improve clinical outcomes for patients with these rare cancers.
© 2022. The Author(s).
Conflict of interest statement
M.M.G.: honoraria from Amgen, Bayer, Epizyme, Daiichi-Sankyo, Flatiron Therapeutics, Karyopharm, Medscape, PER, SpringWorks Therapeutics, Tracon, and UpToDate; D.X.J., S.E.T., J.L., G.M.F., J.M.V., S.Z.M.: employment by Foundation Medicine, Inc. and stock ownership in Roche; S.A.: research funding from AB Science, Adaptimmune, Advenchen Laboratories, Bavarian Nordic, Bayer, Blueprint Medicines, BTG Specialty Pharmaceuticals, CBA Pharma, Deciphera, Eli Lilly, Epizyme, Forma Therapeutics, Genmab, GlaxoSmithKline, Gradalis, Immune Design, Incyte, Karyopharm, Merck, Novartis, Philogen, PTC Therapeutics, SpringWorks Therapeutics, Takeda, TRACON Pharma; W.A.: research funding from AstraZeneca, Clovis Oncology, GlaxoSmithKline, Zenith Epigenetics; honoraria from Caret Pharma; consulting or advisory work for Clovis Oncology, Daiichi Sankyo, Janssen, MORE Health, and ORIC Pharmaceuticals; travel expenses from Clovis Oncology, GlaxoSmithKline, and ORIC Pharmaceuticals; A.D.: honoraria from or advisory work for Abbvie, ArcherDX, ARIAD Pharmaceuticals, AstraZeneca, Axis Therapeutics, Beigene, BergenBio, Blueprint Medicines, Elevation Oncology, Exelixis, Foundation Medicine, Genentech, Helsinn, Hengrui Therapeutics, Ignyta, Loxo Oncology, Medscape, Millennium Pharmaceuticals, MORE Health, OncLive, Paradigm Medical Communications, Peerview Institute, PeerVoice, Pfizer, Physicians Education Resources, Remedica Ltd., Research to Practice, Targeted Oncology, TP Therapeutics, Tyra Biosciences, Verastem, and WebMD; research funding from Exelixis, Foundation Medicine, GlaxoSmithKline, Pfizer, PharmaMar, Teva Pharmaceuticals, and Taiho; royalties from Wolters Kluwer; travel expenses from Boehringer Ingelheim, Merck, Merus, and Puma; P.C.: research funding from Array Biopharma and Novartis; advisory and consulting work for Deciphera and Exelixis; SPD: research funding from Amgen, Bristol-Myers Squibb, Deciphera, EMD Serono, Incyte, Merck, Nektar; consulting or advisory work for Amgen, GlaxoSmithKline, Immune Design, Immunocore, Incyte, Merck, Nektar, Pfizer; travel expenses from Adaptimmune, EMD Serono, Immunocore, Merck; M.A.D.: research funding from Aadi Bioscience and Eli Lilly; C.F.M.: advisory work for Bayer and honoraria from Novartis; S.P.C.: research funding from Amgen, Ignyta, Immune Design, Karyopharm, Roche, Threshold Pharmaceuticals, and the Sarcoma Alliance for Research through Collaboration (SARC); E.C.: research funding from Adaptimmune, Amgen, AstraZeneca, Bayer, Exelixis, GlaxoSmithKline, Iterion Therapeutics, Merck, Mirati, and Novartis; A.J.W.: research funding from Aadi Bioscience, Daiichi-Sankyo, Eli Lilly, Karyopharm, Plexxikon; consulting for Daiichi-Sankyo, Deciphera, Nanocarrier; A.S.S.: research support from Blueprint Medicines, Deciphera, Eli Lilly, and Nanocarrier; consulting or speakers’ bureau fees from Certis Oncology, Daiichi Sankyo, Deciphera, Eisai, Eli Lilly, Novartis, OncLive, and Roche; J.S.R.: employee of Foundation Medicine, Inc., board observer for Celsius Therapeutics, advisory work for Tango Therapeutics, stock ownership in the same 3 companies; D.M.T.: employee of Omico; research funding or support from Amgen, Astra Zeneca, Bayer, Eisai, Merck, Pfizer, and Roche; honoraria from Bayer, Merck, Pfizer, and Roche; RGM: research funding from Bayer, Eli Lilly, Karyopharm, Pfizer, Presage, Regeneron, SARC, SpringWorks Therapeutics; honoraria or consulting work for Bayer, Deciphera, Eisai, Epizyme, Foundation Medicine, GlaxoSmithKline, Imclone, Immune Design, Karyopharm, Novartis, Pfizer, PharmaMar, Presage, SARC, SpringWorks Therapeutics, UptoDate; W.D.T.: advisory board and consulting fees from Agios Pharmaceuticals, Blueprint Medicines, Daiichi Sankyo, Deciphera, Eisai, Eli Lilly, EMD Serono, GlaxoSmithKline, Immune Design, Janssen, and NanoCarrier; advisory board membership with stock interests for Atropos Therapeutics and Certis Oncology Solutions; pending patent through MSK for biomarkers of CDK4 inhibition; S.M.A.: employee of EQRx Inc. and advisor for Incysus Therapeutics and Elevation Oncology. All other authors have no relationships to disclose.
Figures



References
-
- Fletcher, C. D. M., World Health Organization. & International Agency for Research on Cancer. WHO classification of tumours of soft tissue and bone, (IARC Press, Lyon, 2013).
-
- Brennan, M. F., Antonescu, C. R., Alektiar, K. & Maki, R. G. Management of soft tissue sarcoma, (Springer, New York, NY, 2016).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

