Identification of miRNAs of Strongyloides stercoralis L1 and iL3 larvae isolated from human stool
- PMID: 35705621
- PMCID: PMC9200769
- DOI: 10.1038/s41598-022-14185-y
Identification of miRNAs of Strongyloides stercoralis L1 and iL3 larvae isolated from human stool
Abstract
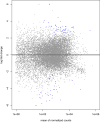
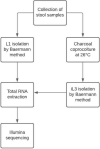
Strongyloidiasis is a neglected tropical disease caused by the soil-transmitted nematode by Strongyloides stercoralis, that affects approximately 600 million people worldwide. In immunosuppressed individuals disseminated strongyloidiasis can rapidly lead to fatal outcomes. There is no gold standard for diagnosing strongyloidiasis, and infections are frequently misdiagnosed. A better understanding of the molecular biology of this parasite can be useful for example for the discovery of potential new biomarkers. Interestingly, recent evidence showed the presence of small RNAs in Strongyloididae, but no data was provided for S. stercoralis. In this study, we present the first identification of miRNAs of both L1 and iL3 larval stages of S. stercoralis. For our purpose, the aims were: (i) to analyse the miRNome of L1 and iL3 S. stercoralis and to identify potential miRNAs of this nematode, (ii) to obtain the mRNAs profiles in these two larval stages and (iii) to predict potential miRNA target sites in mRNA sequences. Total RNA was isolated from L1 and iL3 collected from the stool of 5 infected individuals. For the miRNAs analysis, we used miRDeep2 software and a pipeline of bio-informatic tools to construct a catalog of a total of 385 sequences. Among these, 53% were common to S. ratti, 19% to S. papillosus, 1% to Caenorhabditis elegans and 44% were novel. Using a differential analysis between the larval stages, we observed 6 suggestive modulated miRNAs (STR-MIR-34A-3P, STR-MIR-8397-3P, STR-MIR-34B-3P and STR-MIR-34C-3P expressed more in iL3, and STR-MIR-7880H-5P and STR-MIR-7880M-5P expressed more in L1). Along with this analysis, we obtained also the mRNAs profiles in the same samples of larvae. Multiple testing found 81 statistically significant mRNAs of the total 1553 obtained (FDR < 0.05; 32 genes expressed more in L1 than iL3; 49 genes expressed more in L3 than iL1). Finally, we found 33 predicted mRNA targets of the modulated miRNAs, providing relevant data for a further validation to better understand the role of these small molecules in the larval stages and their valuein clinical diagnostics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Novel insights into the somatic proteome of Strongyloides stercoralis infective third-stage larvae.Parasit Vectors. 2023 Jan 31;16(1):45. doi: 10.1186/s13071-023-05675-7. Parasit Vectors. 2023. PMID: 36721249 Free PMC article.
-
Diagnosis of Strongyloides stercoralis by morphological characteristics combine with molecular biological methods.Parasitol Res. 2017 Apr;116(4):1159-1163. doi: 10.1007/s00436-017-5389-y. Epub 2017 Jan 26. Parasitol Res. 2017. PMID: 28124134
-
Development and Efficacy of Droplet Digital PCR for Detection of Strongyloides stercoralis in Stool.Am J Trop Med Hyg. 2021 Oct 18;106(1):312-319. doi: 10.4269/ajtmh.21-0729. Am J Trop Med Hyg. 2021. PMID: 34662861 Free PMC article.
-
From past to present: opportunities and trends in the molecular detection and diagnosis of Strongyloides stercoralis.Parasit Vectors. 2023 Apr 11;16(1):123. doi: 10.1186/s13071-023-05763-8. Parasit Vectors. 2023. PMID: 37041645 Free PMC article. Review.
-
[An overview of Strongyloides stercoralis and its infections].Mikrobiyol Bul. 2009 Jan;43(1):169-77. Mikrobiyol Bul. 2009. PMID: 19334396 Review. Turkish.
Cited by
-
Advancing Strongyloides omics data: bridging the gap with Caenorhabditis elegans.Philos Trans R Soc Lond B Biol Sci. 2024 Jan 15;379(1894):20220437. doi: 10.1098/rstb.2022.0437. Epub 2023 Nov 27. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38008117 Free PMC article. Review.
-
Novel insights into the somatic proteome of Strongyloides stercoralis infective third-stage larvae.Parasit Vectors. 2023 Jan 31;16(1):45. doi: 10.1186/s13071-023-05675-7. Parasit Vectors. 2023. PMID: 36721249 Free PMC article.
-
Present status with impacts and roles of miRNA on Soil Transmitted Helminthiosis control: A review.Curr Res Pharmacol Drug Discov. 2023 Jul 15;5:100162. doi: 10.1016/j.crphar.2023.100162. eCollection 2023. Curr Res Pharmacol Drug Discov. 2023. PMID: 37520661 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

