A method to investigate muscle target-specific transcriptional signatures of single motor neurons
- PMID: 35705847
- PMCID: PMC10084336
- DOI: 10.1002/dvdy.507
A method to investigate muscle target-specific transcriptional signatures of single motor neurons
Abstract
Background: Motor neurons in the vertebrate spinal cord have long served as a paradigm to study the transcriptional logic of cell type specification and differentiation. At limb levels, pool-specific transcriptional signatures first restrict innervation to only one particular muscle in the periphery, and get refined, once muscle connection has been established. Accordingly, to study the transcriptional dynamics and specificity of the system, a method for establishing muscle target-specific motor neuron transcriptomes would be required.
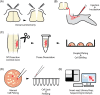
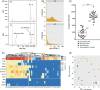
Results: To investigate target-specific transcriptional signatures of single motor neurons, here we combine ex-ovo retrograde axonal labeling in mid-gestation chicken embryos with manual isolation of individual fluorescent cells and Smart-seq2 single-cell RNA-sequencing. We validate our method by injecting the dorsal extensor metacarpi radialis and ventral flexor digiti quarti wing muscles and harvesting a total of 50 fluorescently labeled cells, in which we detect up to 12,000 transcribed genes. Additionally, we present visual cues and cDNA metrics predictive of sequencing success.
Conclusions: Our method provides a unique approach to study muscle target-specific motor neuron transcriptomes at a single-cell resolution. We anticipate that our method will provide key insights into the transcriptional logic underlying motor neuron pool specialization and proper neuromuscular circuit assembly and refinement.
Keywords: Smart-seq2; axonal backfill; limb motor neuron-muscle connection; manual cell picking; neural tube dissociation; single motor neuron sequencing.
© 2022 The Authors. Developmental Dynamics published by Wiley Periodicals LLC on behalf of American Association for Anatomy.
Figures


Similar articles
-
Effect of sonic hedgehog on motor neuron positioning in the spinal cord during chicken embryonic development.J Cell Mol Med. 2019 May;23(5):3549-3562. doi: 10.1111/jcmm.14254. Epub 2019 Mar 4. J Cell Mol Med. 2019. PMID: 30834718 Free PMC article.
-
A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity.Cell. 2005 Nov 4;123(3):477-91. doi: 10.1016/j.cell.2005.09.009. Cell. 2005. PMID: 16269338
-
Wnt3a Ectopic Expression Interferes Axonal Projection and Motor Neuron Positioning During the Chicken Spinal Cord Development.J Mol Neurosci. 2018 Apr;64(4):619-630. doi: 10.1007/s12031-018-1060-z. Epub 2018 Mar 25. J Mol Neurosci. 2018. PMID: 29574664
-
Axon pathfinding for locomotion.Semin Cell Dev Biol. 2019 Jan;85:26-35. doi: 10.1016/j.semcdb.2017.11.014. Epub 2017 Nov 14. Semin Cell Dev Biol. 2019. PMID: 29141181 Review.
-
Transcriptional codes and the control of neuronal identity.Annu Rev Neurosci. 2002;25:251-81. doi: 10.1146/annurev.neuro.25.112701.142916. Epub 2002 Mar 27. Annu Rev Neurosci. 2002. PMID: 12052910 Review.
Cited by
-
Distinct patterning responses of wing and leg neuromuscular systems to different preaxial polydactylies.Front Cell Dev Biol. 2023 May 4;11:1154205. doi: 10.3389/fcell.2023.1154205. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37215090 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

