Silencing the ADAM9 Gene through CRISPR/Cas9 Protects Mice from Alcohol-Induced Acute Liver Injury
- PMID: 35707386
- PMCID: PMC9192226
- DOI: 10.1155/2022/5110161
Silencing the ADAM9 Gene through CRISPR/Cas9 Protects Mice from Alcohol-Induced Acute Liver Injury
Abstract
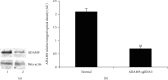
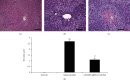
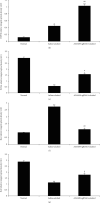
Alcoholic liver injury is a major global public health concern at present. The ADAM9 gene plays a crucial role in the occurrence and development of various liver diseases, but its role in acute alcoholic liver injury remains ambiguous. In this study, a chimeric single-guide RNA targeting the genomic regions of mouse ADAM9 was designed using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology. Next, the role of ADAM9 in acute alcoholic liver injury in vitro in cultured mouse cells and in vivo in a hydrodynamic injection-based alcoholic liver injury mouse model was documented. The findings of this study suggest that ADAM9 induces by regulating cell proliferation, apoptosis, and stress metabolism in mice. Thus, inhibiting the expression of ADAM9 gene using CRISPR/Cas9 can attenuate alcohol-induced acute liver injury in mice.
Copyright © 2022 Yong-Yong Zhang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures







References
-
- Zhang Y.-Y., Li S.-Q., Song Y., et al. The therapeutic effect of Chaihu-Shugan-San in fatty liver disease: a meta-analysis randomized controlled trials. International Journal of Clinical and Experimental Medicine . 2018;11:12880–12888.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

