Metalloproteinase-Dependent and TMPRSS2-Independent Cell Surface Entry Pathway of SARS-CoV-2 Requires the Furin Cleavage Site and the S2 Domain of Spike Protein
- PMID: 35708281
- PMCID: PMC9426510
- DOI: 10.1128/mbio.00519-22
Metalloproteinase-Dependent and TMPRSS2-Independent Cell Surface Entry Pathway of SARS-CoV-2 Requires the Furin Cleavage Site and the S2 Domain of Spike Protein
Abstract
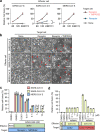
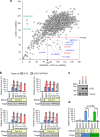
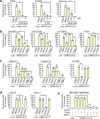
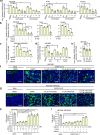
The ongoing global vaccination program to prevent SARS-CoV-2 infection, the causative agent of COVID-19, has had significant success. However, recently, virus variants that can evade the immunity in a host achieved through vaccination have emerged. Consequently, new therapeutic agents that can efficiently prevent infection from these new variants, and hence COVID-19 spread, are urgently required. To achieve this, extensive characterization of virus-host cell interactions to identify effective therapeutic targets is warranted. Here, we report a cell surface entry pathway of SARS-CoV-2 that exists in a cell type-dependent manner and is TMPRSS2 independent but sensitive to various broad-spectrum metalloproteinase inhibitors such as marimastat and prinomastat. Experiments with selective metalloproteinase inhibitors and gene-specific small interfering RNAS (siRNAs) revealed that a disintegrin and metalloproteinase 10 (ADAM10) is partially involved in the metalloproteinase pathway. Consistent with our finding that the pathway is unique to SARS-CoV-2 among highly pathogenic human coronaviruses, both the furin cleavage motif in the S1/S2 boundary and the S2 domain of SARS-CoV-2 spike protein are essential for metalloproteinase-dependent entry. In contrast, the two elements of SARS-CoV-2 independently contributed to TMPRSS2-dependent S2 priming. The metalloproteinase pathway is involved in SARS-CoV-2-induced syncytium formation and cytopathicity, leading us to theorize that it is also involved in the rapid spread of SARS-CoV-2 and the pathogenesis of COVID-19. Thus, targeting the metalloproteinase pathway in addition to the TMPRSS2 and endosomal pathways could be an effective strategy by which to cure COVID-19 in the future. IMPORTANCE To develop effective therapeutics against COVID-19, it is necessary to elucidate in detail the infection mechanism of the causative agent, SARS-CoV-2. SARS-CoV-2 binds to the cell surface receptor ACE2 via the spike protein, and then the spike protein is cleaved by host proteases to enable entry. Here, we found that the metalloproteinase-mediated pathway is important for SARS-CoV-2 infection in addition to the TMPRSS2-mediated pathway and the endosomal pathway. The metalloproteinase-mediated pathway requires both the prior cleavage of spike into two domains and a specific sequence in the second domain, S2, conditions met by SARS-CoV-2 but lacking in the related human coronavirus SARS-CoV. Besides the contribution of metalloproteinases to SARS-CoV-2 infection, inhibition of metalloproteinases was important in preventing cell death, which may cause organ damage. Our study provides new insights into the complex pathogenesis unique to COVID-19 and relevant to the development of effective therapies.
Keywords: SARS-CoV-2; furin; membrane fusion; metalloproteinase; virus entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity.J Virol. 2022 Apr 27;96(8):e0012822. doi: 10.1128/jvi.00128-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343766 Free PMC article.
-
SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation.J Virol. 2021 Apr 12;95(9):e00002-21. doi: 10.1128/JVI.00002-21. Print 2021 Apr 12. J Virol. 2021. PMID: 33608407 Free PMC article.
-
SARS-CoV-2 Spike Furin Cleavage Site and S2' Basic Residues Modulate the Entry Process in a Host Cell-Dependent Manner.J Virol. 2022 Jul 13;96(13):e0047422. doi: 10.1128/jvi.00474-22. Epub 2022 Jun 9. J Virol. 2022. PMID: 35678602 Free PMC article.
-
Proteolytic activation of SARS-CoV-2 spike protein.Microbiol Immunol. 2022 Jan;66(1):15-23. doi: 10.1111/1348-0421.12945. Epub 2021 Oct 12. Microbiol Immunol. 2022. PMID: 34561887 Free PMC article. Review.
-
Targeting the viral-entry facilitators of SARS-CoV-2 as a therapeutic strategy in COVID-19.J Med Virol. 2021 Sep;93(9):5260-5276. doi: 10.1002/jmv.27019. Epub 2021 May 3. J Med Virol. 2021. PMID: 33851732 Free PMC article. Review.
Cited by
-
Identification and differential usage of a host metalloproteinase entry pathway by SARS-CoV-2 Delta and Omicron.iScience. 2022 Nov 18;25(11):105316. doi: 10.1016/j.isci.2022.105316. Epub 2022 Oct 10. iScience. 2022. PMID: 36254158 Free PMC article.
-
iNGNN-DTI: prediction of drug-target interaction with interpretable nested graph neural network and pretrained molecule models.Bioinformatics. 2024 Mar 4;40(3):btae135. doi: 10.1093/bioinformatics/btae135. Bioinformatics. 2024. PMID: 38449285 Free PMC article.
-
Potential Effects of Hyperglycemia on SARS-CoV-2 Entry Mechanisms in Pancreatic Beta Cells.Viruses. 2024 Aug 2;16(8):1243. doi: 10.3390/v16081243. Viruses. 2024. PMID: 39205219 Free PMC article. Review.
-
Metalloprotease-Dependent S2'-Activation Promotes Cell-Cell Fusion and Syncytiation of SARS-CoV-2.Viruses. 2022 Sep 21;14(10):2094. doi: 10.3390/v14102094. Viruses. 2022. PMID: 36298651 Free PMC article.
-
SP-A binding to the SARS-CoV-2 spike protein using hybrid quantum and classical in silico modeling and molecular pruning by Quantum Approximate Optimization Algorithm (QAOA) Based MaxCut with ZDOCK.Front Immunol. 2022 Sep 13;13:945317. doi: 10.3389/fimmu.2022.945317. eCollection 2022. Front Immunol. 2022. PMID: 36189278 Free PMC article.
References
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris JSM, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 362:1353–1358. doi:10.1016/S0140-6736(03)14630-2. - DOI - PMC - PubMed
-
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguière AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi:10.1056/NEJMoa030747. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

