Genomic Analysis of Carbapenem-Resistant Comamonas in Water Matrices: Implications for Public Health and Wastewater Treatments
- PMID: 35708324
- PMCID: PMC9275215
- DOI: 10.1128/aem.00646-22
Genomic Analysis of Carbapenem-Resistant Comamonas in Water Matrices: Implications for Public Health and Wastewater Treatments
Abstract
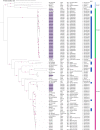
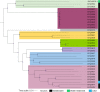
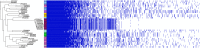
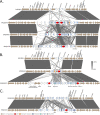
Comamonas spp. are Gram-negative bacteria that catabolize a wide range of organic and inorganic substrates. Comamonas spp. are abundant in aquatic and soil environments, including wastewater, and can cause opportunistic infections in humans. Because of their potential in wastewater bioaugmentation and bioremediation strategies, the identification of Comamonas species harboring genes encoding carbapenemases and other clinically important antibiotic resistance genes warrant further investigation. Here, we present an analysis of 39 whole-genome sequences comprising three Comamonas species from aquatic environments in South Australia that were recovered on media supplemented with carbapenems. The analysis includes a detailed description of 33 Comamonas denitrificans isolates, some of which carried chromosomally acquired blaGES-5, blaOXA, and aminoglycoside resistance (aadA) genes located on putative genomic islands (GIs). All blaGES-5- and blaOXA-containing GIs appear to be unique to this Australian collection of C. denitrificans. Notably, most open reading frames (ORFs) within the GIs, including all antimicrobial resistance (AMR) genes, had adjacent attC sites, indicating that these ORFs are mobile gene cassettes. One C. denitrificans isolate carried an IncP-1 plasmid with genes involved in xenobiotic degradation and response to oxidative stress. Our assessment of the sequences highlights the very distant nature of C. denitrificans to the other Comamonas species and its apparent disposition to acquire antimicrobial resistance genes on putative genomic islands. IMPORTANCE Antimicrobial resistance (AMR) poses a global public health threat, and the increase in resistance to "last-resort drugs," such as carbapenems, is alarming. Wastewater has been flagged as a hot spot for AMR evolution. Comamonas spp. are among the most common bacteria in wastewater and play a role in its bioaugmentation. While the ability of Comamonas species to catabolize a wide range of organic and inorganic substrates is well documented, some species are also opportunistic pathogens. However, data regarding AMR in Comamonas spp. are limited. Here, through the genomic analyses of 39 carbapenem-resistant Comamonas isolates, we make several key observations, including the identification of a subset of C. denitrificans isolates that harbored genomic islands encoding carbapenemase blaGES-5 or extended-spectrum β-lactamase blaOXA alleles. Given the importance of Comamonas species in potential wastewater bioaugmentation and bioremediation strategies, as well as their status as emerging pathogens, the acquisition of critically important antibiotic resistance genes on genomic islands warrants future monitoring.
Keywords: Comamonas; Comamonas denitrificans; antimicrobial resistance (AMR); blaGES-5; carbapenemase; genomic islands; incP-1; wastewater; whole-genome sequencing (WGS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

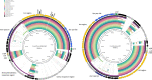
References
-
- Brady MT, Marcon MJ. 2008. Less commonly encountered nonenteric Gram-negative bacilli, p 828–831. In Long SS (ed), Principles and Practice of Pediatric Infectious Disease, 3rd ed. W.B. Saunders, Philadelphia, PA. 10.1016/B978-0-7020-3468-8.50157-7. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

