Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia
- PMID: 35708626
- PMCID: PMC9206114
- DOI: 10.1084/jem.20220131
Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia
Abstract
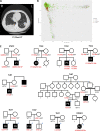
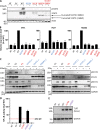
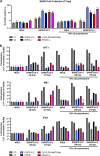
Recessive or dominant inborn errors of type I interferon (IFN) immunity can underlie critical COVID-19 pneumonia in unvaccinated adults. The risk of COVID-19 pneumonia in unvaccinated children, which is much lower than in unvaccinated adults, remains unexplained. In an international cohort of 112 children (<16 yr old) hospitalized for COVID-19 pneumonia, we report 12 children (10.7%) aged 1.5-13 yr with critical (7 children), severe (3), and moderate (2) pneumonia and 4 of the 15 known clinically recessive and biochemically complete inborn errors of type I IFN immunity: X-linked recessive TLR7 deficiency (7 children) and autosomal recessive IFNAR1 (1), STAT2 (1), or TYK2 (3) deficiencies. Fibroblasts deficient for IFNAR1, STAT2, or TYK2 are highly vulnerable to SARS-CoV-2. These 15 deficiencies were not found in 1,224 children and adults with benign SARS-CoV-2 infection without pneumonia (P = 1.2 × 10-11) and with overlapping age, sex, consanguinity, and ethnicity characteristics. Recessive complete deficiencies of type I IFN immunity may underlie ∼10% of hospitalizations for COVID-19 pneumonia in children.
© 2022 ZHANG et al.
Conflict of interest statement
Disclosures: B.E. Young reported personal fees from Sanofi, Gilead, Roche, Astra-Zeneca, and Novacyte outside the submitted work. R.P. Lifton is a member of the board of directors of Roche and its subsidiary Genentech. I. Meyts reported grants from CSL-Behring outside the submitted work. J.-L. Casanova reported a patent to PCT/US2021/042741 pending. No other disclosures were reported.
Figures

References
-
- Abolhassani, H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., Aranda-Guillen M., Sardh F., Zuo F., Zhang P., et al. 2022. Inherited IFNAR1 deficiency in a child with both critical COVID-19 pneumonia and multisystem inflammatory syndrome. J. Clin. Immunol. 42:471–483. 10.1007/s10875-022-01215-7 - DOI - PMC - PubMed
-
- Abolhassani, H., Vosughimotlagh A., Asano T., Landegren N., Boisson B., Delavari S., Bastard P., Aranda-Guillen M., Wang Y., Zuo F., et al. 2021. X-linked TLR7 deficiency underlies critical COVID-19 pneumonia in a male patient with Ataxia-Telangiectasia. J. Clin. Immunol. 42:1–9. 10.1007/s10875-021-01151-y - DOI - PMC - PubMed
-
- Asano, T., Boisson B., Onodi F., Matuozzo D., Moncada-Velez M., Maglorius Renkilaraj M.R.L., Zhang P., Meertens L., Bolze A., Materna M., et al. 2021. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 6:eabl4348. 10.1126/sciimmunol.abl4348 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UM1 HG006504/HG/NHGRI NIH HHS/United States
- ERC_/European Research Council/International
- S10 OD018521/OD/NIH HHS/United States
- P01 AI138938/AI/NIAID NIH HHS/United States
- R01 AI088364/AI/NIAID NIH HHS/United States
- U19 AI111825/AI/NIAID NIH HHS/United States
- R01 AI163029/AI/NIAID NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- MR/S032304/1/MRC_/Medical Research Council/United Kingdom
- R01 AI091707/AI/NIAID NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- R01 AI161444/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UL1 RR024143/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

