The retrograde signaling regulator ANAC017 recruits the MKK9-MPK3/6, ethylene, and auxin signaling pathways to balance mitochondrial dysfunction with growth
- PMID: 35708648
- PMCID: PMC9421482
- DOI: 10.1093/plcell/koac177
The retrograde signaling regulator ANAC017 recruits the MKK9-MPK3/6, ethylene, and auxin signaling pathways to balance mitochondrial dysfunction with growth
Abstract
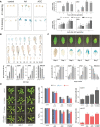
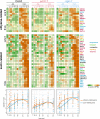
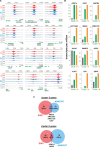
In plant cells, mitochondria are ideally positioned to sense and balance changes in energy metabolism in response to changing environmental conditions. Retrograde signaling from mitochondria to the nucleus is crucial for adjusting the required transcriptional responses. We show that ANAC017, the master regulator of mitochondrial stress, directly recruits a signaling cascade involving the plant hormones ethylene and auxin as well as the MAP KINASE KINASE (MKK) 9-MAP KINASE (MPK) 3/6 pathway in Arabidopsis thaliana. Chromatin immunoprecipitation followed by sequencing and overexpression demonstrated that ANAC017 directly regulates several genes of the ethylene and auxin pathways, including MKK9, 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, and YUCCA 5, in addition to genes encoding transcription factors regulating plant growth and stress responses such as BASIC REGION/LEUCINE ZIPPER MOTIF (bZIP) 60, bZIP53, ANAC081/ATAF2, and RADICAL-INDUCED CELL DEATH1. A time-resolved RNA-seq experiment established that ethylene signaling precedes the stimulation of auxin signaling in the mitochondrial stress response, with a large part of the transcriptional regulation dependent on ETHYLENE-INSENSITIVE 3. These results were confirmed by mutant analyses. Our findings identify the molecular components controlled by ANAC017, which integrates the primary stress responses to mitochondrial dysfunction with whole plant growth via the activation of regulatory and partly antagonistic feedback loops.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures




Similar articles
-
ANAC017 Coordinates Organellar Functions and Stress Responses by Reprogramming Retrograde Signaling.Plant Physiol. 2019 May;180(1):634-653. doi: 10.1104/pp.18.01603. Epub 2019 Mar 14. Plant Physiol. 2019. PMID: 30872424 Free PMC article.
-
Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance.Plant J. 2016 Nov;88(4):542-558. doi: 10.1111/tpj.13276. Epub 2016 Oct 17. Plant J. 2016. PMID: 27425258
-
Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis.J Biol Chem. 2008 Oct 3;283(40):26996-7006. doi: 10.1074/jbc.M801392200. Epub 2008 Aug 7. J Biol Chem. 2008. PMID: 18693252
-
Deciphering Auxin-Ethylene Crosstalk at a Systems Level.Int J Mol Sci. 2018 Dec 14;19(12):4060. doi: 10.3390/ijms19124060. Int J Mol Sci. 2018. PMID: 30558241 Free PMC article. Review.
-
Paradigms and paradox in the ethylene signaling pathway and interaction network.Mol Plant. 2011 Jul;4(4):626-34. doi: 10.1093/mp/ssr042. Epub 2011 Jun 20. Mol Plant. 2011. PMID: 21690206 Review.
Cited by
-
Genome-Wide Identification of MKK Gene Family and Response to Hormone and Abiotic Stress in Rice.Plants (Basel). 2024 Oct 18;13(20):2922. doi: 10.3390/plants13202922. Plants (Basel). 2024. PMID: 39458871 Free PMC article.
-
Molecular traits of MAPK kinases and the regulatory mechanism of GhMAPKK5 alleviating drought/salt stress in cotton.Plant Physiol. 2024 Nov 4;196(3):2030-2047. doi: 10.1093/plphys/kiae415. Plant Physiol. 2024. PMID: 39140753 Free PMC article.
-
Perception and processing of stress signals by plant mitochondria.Plant J. 2024 Dec;120(6):2337-2355. doi: 10.1111/tpj.17133. Epub 2024 Nov 11. Plant J. 2024. PMID: 39527570 Free PMC article. Review.
-
Nuclear pyruvate dehydrogenase complex regulates histone acetylation and transcriptional regulation in the ethylene response.Sci Adv. 2024 Jul 26;10(30):eado2825. doi: 10.1126/sciadv.ado2825. Epub 2024 Jul 26. Sci Adv. 2024. PMID: 39058774 Free PMC article.
-
ANAC044 orchestrates mitochondrial stress signaling to trigger iron-induced stem cell death in root meristems.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2411579122. doi: 10.1073/pnas.2411579122. Epub 2024 Dec 30. Proc Natl Acad Sci U S A. 2025. PMID: 39793035 Free PMC article.
References
-
- Berkowitz O, Xu Y, Liew LC, Wang Y, Zhu Y, Hurgobin B, Lewsey MG, Whelan J (2021) RNA-seq analysis of laser microdissected Arabidopsis thaliana leaf epidermis, mesophyll and vasculature defines tissue-specific transcriptional responses to multiple stress treatments. Plant J 107: 938–955 - PubMed
-
- Braam J (2005) In touch: plant responses to mechanical stimuli. New Phytol 165: 373–389 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

