Infection of lung megakaryocytes and platelets by SARS-CoV-2 anticipate fatal COVID-19
- PMID: 35708858
- PMCID: PMC9201269
- DOI: 10.1007/s00018-022-04318-x
Infection of lung megakaryocytes and platelets by SARS-CoV-2 anticipate fatal COVID-19
Abstract
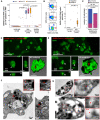
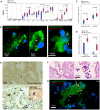
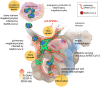
SARS-CoV-2, although not being a circulatory virus, spread from the respiratory tract resulting in multiorgan failures and thrombotic complications, the hallmarks of fatal COVID-19. A convergent contributor could be platelets that beyond hemostatic functions can carry infectious viruses. Here, we profiled 52 patients with severe COVID-19 and demonstrated that circulating platelets of 19 out 20 non-survivor patients contain SARS-CoV-2 in robust correlation with fatal outcome. Platelets containing SARS-CoV-2 might originate from bone marrow and lung megakaryocytes (MKs), the platelet precursors, which were found infected by SARS-CoV-2 in COVID-19 autopsies. Accordingly, MKs undergoing shortened differentiation and expressing anti-viral IFITM1 and IFITM3 RNA as a sign of viral sensing were enriched in the circulation of deadly COVID-19. Infected MKs reach the lung concomitant with a specific MK-related cytokine storm rich in VEGF, PDGF and inflammatory molecules, anticipating fatal outcome. Lung macrophages capture SARS-CoV-2-containing platelets in vivo. The virus contained by platelets is infectious as capture of platelets carrying SARS-CoV-2 propagates infection to macrophages in vitro, in a process blocked by an anti-GPIIbIIIa drug. Altogether, platelets containing infectious SARS-CoV-2 alter COVID-19 pathogenesis and provide a powerful fatality marker. Clinical targeting of platelets might prevent viral spread, thrombus formation and exacerbated inflammation at once and increase survival in COVID-19.
Keywords: COVID-19; Lung; Macrophages; Megakaryocytes; Platelets; SARS-CoV-2.
© 2022. The Author(s).
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures


Similar articles
-
Cardiac megakaryocytes in SARS-CoV-2-positive autopsies.Histopathology. 2022 Nov;81(5):600-624. doi: 10.1111/his.14734. Epub 2022 Sep 7. Histopathology. 2022. PMID: 35925828 Free PMC article.
-
Characterization of all small RNAs in and comparisons across cultured megakaryocytes and platelets of healthy individuals and COVID-19 patients.J Thromb Haemost. 2023 Nov;21(11):3252-3267. doi: 10.1016/j.jtha.2023.07.028. Epub 2023 Aug 7. J Thromb Haemost. 2023. PMID: 37558133
-
Circulating SARS-CoV-2+ megakaryocytes are associated with severe viral infection in COVID-19.Blood Adv. 2023 Aug 8;7(15):4200-4214. doi: 10.1182/bloodadvances.2022009022. Blood Adv. 2023. PMID: 36920790 Free PMC article.
-
Platelets and COVID-19.Hamostaseologie. 2021 Oct;41(5):379-385. doi: 10.1055/a-1581-4355. Epub 2021 Oct 25. Hamostaseologie. 2021. PMID: 34695854 Review.
-
Occurrence and role of lung megakaryocytes in infection and inflammation.Front Immunol. 2022 Nov 29;13:1029223. doi: 10.3389/fimmu.2022.1029223. eCollection 2022. Front Immunol. 2022. PMID: 36524131 Free PMC article. Review.
Cited by
-
An Autopsy Case of Severe COVID-19 Pneumonia Complicated by Intrapulmonary Thrombosis in Myelodysplastic/Myeloproliferative Neoplasm With Ring Sideroblasts and Thrombocytosis.Cureus. 2024 Jun 20;16(6):e62790. doi: 10.7759/cureus.62790. eCollection 2024 Jun. Cureus. 2024. PMID: 39036222 Free PMC article.
-
Research progress of megakaryocytes and platelets in lung injury.Ann Med. 2024 Dec;56(1):2362871. doi: 10.1080/07853890.2024.2362871. Epub 2024 Jun 20. Ann Med. 2024. PMID: 38902986 Free PMC article. Review.
-
HIV-1 inhibits IFITM3 expression to promote the infection of megakaryocytes.J Mol Cell Biol. 2025 Mar 21;16(9):mjae042. doi: 10.1093/jmcb/mjae042. J Mol Cell Biol. 2025. PMID: 39354676 Free PMC article.
-
Breakthrough infections after COVID-19 vaccinations do not elicit platelet hyperactivation and are associated with high platelet-lymphocyte and low platelet-neutrophil aggregates.Res Pract Thromb Haemost. 2023 Nov 14;7(8):102262. doi: 10.1016/j.rpth.2023.102262. eCollection 2023 Nov. Res Pract Thromb Haemost. 2023. PMID: 38193050 Free PMC article.
-
Persistent but dysfunctional mucosal SARS-CoV-2-specific IgA and low lung IL-1β associate with COVID-19 fatal outcome: A cross-sectional analysis.Front Immunol. 2022 Sep 29;13:842468. doi: 10.3389/fimmu.2022.842468. eCollection 2022. Front Immunol. 2022. PMID: 36248831 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

