Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon
- PMID: 35709277
- PMCID: PMC10064918
- DOI: 10.1126/science.abg3875
Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon
Abstract
The elongation of eukaryotic selenoproteins relies on a poorly understood process of interpreting in-frame UGA stop codons as selenocysteine (Sec). We used cryo-electron microscopy to visualize Sec UGA recoding in mammals. A complex between the noncoding Sec-insertion sequence (SECIS), SECIS-binding protein 2 (SBP2), and 40S ribosomal subunit enables Sec-specific elongation factor eEFSec to deliver Sec. eEFSec and SBP2 do not interact directly but rather deploy their carboxyl-terminal domains to engage with the opposite ends of the SECIS. By using its Lys-rich and carboxyl-terminal segments, the ribosomal protein eS31 simultaneously interacts with Sec-specific transfer RNA (tRNASec) and SBP2, which further stabilizes the assembly. eEFSec is indiscriminate toward l-serine and facilitates its misincorporation at Sec UGA codons. Our results support a fundamentally distinct mechanism of Sec UGA recoding in eukaryotes from that in bacteria.
Conflict of interest statement
Figures

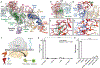


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

