Single-cell multiomics reveal the scale of multilayered adaptations enabling CLL relapse during venetoclax therapy
- PMID: 35709339
- PMCID: PMC10653037
- DOI: 10.1182/blood.2022016040
Single-cell multiomics reveal the scale of multilayered adaptations enabling CLL relapse during venetoclax therapy
Abstract
Venetoclax (VEN) inhibits the prosurvival protein BCL2 to induce apoptosis and is a standard therapy for chronic lymphocytic leukemia (CLL), delivering high complete remission rates and prolonged progression-free survival in relapsed CLL but with eventual loss of efficacy. A spectrum of subclonal genetic changes associated with VEN resistance has now been described. To fully understand clinical resistance to VEN, we combined single-cell short- and long-read RNA-sequencing to reveal the previously unappreciated scale of genetic and epigenetic changes underpinning acquired VEN resistance. These appear to be multilayered. One layer comprises changes in the BCL2 family of apoptosis regulators, especially the prosurvival family members. This includes previously described mutations in BCL2 and amplification of the MCL1 gene but is heterogeneous across and within individual patient leukemias. Changes in the proapoptotic genes are notably uncommon, except for single cases with subclonal losses of BAX or NOXA. Much more prominent was universal MCL1 gene upregulation. This was driven by an overlying layer of emergent NF-κB (nuclear factor kappa B) activation, which persisted in circulating cells during VEN therapy. We discovered that MCL1 could be a direct transcriptional target of NF-κB. Both the switch to alternative prosurvival factors and NF-κB activation largely dissipate following VEN discontinuation. Our studies reveal the extent of plasticity of CLL cells in their ability to evade VEN-induced apoptosis. Importantly, these findings pinpoint new approaches to circumvent VEN resistance and provide a specific biological justification for the strategy of VEN discontinuation once a maximal response is achieved rather than maintaining long-term selective pressure with the drug.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: R.T., L.T., M.A.A., C.F., A.J., A.L.G., J.S.J., H.P., C.E.T., Q.G., A.G., T.T., T.M.D., D.H.D.G., I.J.M., M.E.R., A.W.R., and D.C.S.H. are employees of the Walter and Eliza Hall Institute, which receives milestone and royalty payments related to venetoclax. M.A.A. has received honorarium from AbbVie. C.S.T. receives research funding and honorarium from both AbbVie and Roche. J.F.S. has received research funding from AbbVie and Genentech and is a consultant and member of advisory boards for both companies. D.H.D.G. has received research funding from Servier. A.W.R. has received research funding from AbbVie. The remaining authors declare no competing financial interests.
The current affiliation for L.T. is the Broad Institute of Harvard and the Massachusetts Institute of Technology, Cambridge, MA.
The current affiliation for T.M.D. is the Hudson Institute of Medical Research, Melbourne, Australia.
The current affiliation for C.S.T. is the Alfred Hospital, Melbourne, Australia.
Figures


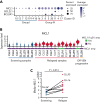
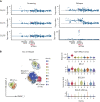



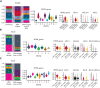

Comment in
-
Mechanisms of resistance to venetoclax.Blood. 2022 Nov 17;140(20):2094-2096. doi: 10.1182/blood.2022017341. Blood. 2022. PMID: 36394906 No abstract available.
References
-
- Roberts AW, Wei AH, Huang DCS. BCL2 and MCL1 inhibitors for hematologic malignancies. Blood. 2021;138(13):1120–1136. - PubMed
-
- Diepstraten ST, Anderson MA, Czabotar PE, Lessene G, Strasser A, Kelly GL. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat Rev Cancer. 2022;22(1):45–64. - PubMed
-
- Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202–208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

