Aberrant EVI1 splicing contributes to EVI1-rearranged leukemia
- PMID: 35709354
- PMCID: PMC9412007
- DOI: 10.1182/blood.2021015325
Aberrant EVI1 splicing contributes to EVI1-rearranged leukemia
Abstract
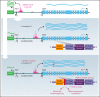
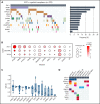
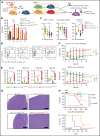
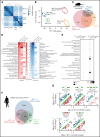
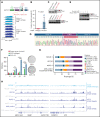
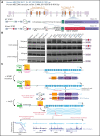
Detailed genomic and epigenomic analyses of MECOM (the MDS1 and EVI1 complex locus) have revealed that inversion or translocation of chromosome 3 drives inv(3)/t(3;3) myeloid leukemias via structural rearrangement of an enhancer that upregulates transcription of EVI1. Here, we identify a novel, previously unannotated oncogenic RNA-splicing derived isoform of EVI1 that is frequently present in inv(3)/t(3;3) acute myeloid leukemia (AML) and directly contributes to leukemic transformation. This EVI1 isoform is generated by oncogenic mutations in the core RNA splicing factor SF3B1, which is mutated in >30% of inv(3)/t(3;3) myeloid neoplasm patients and thereby represents the single most commonly cooccurring genomic alteration in inv(3)/t(3;3) patients. SF3B1 mutations are statistically uniquely enriched in inv(3)/t(3;3) myeloid neoplasm patients and patient-derived cell lines compared with other forms of AML and promote mis-splicing of EVI1 generating an in-frame insertion of 6 amino acids at the 3' end of the second zinc finger domain of EVI1. Expression of this EVI1 splice variant enhanced the self-renewal of hematopoietic stem cells, and introduction of mutant SF3B1 in mice bearing the humanized inv(3)(q21q26) allele resulted in generation of this novel EVI1 isoform in mice and hastened leukemogenesis in vivo. The mutant SF3B1 spliceosome depends upon an exonic splicing enhancer within EVI1 exon 13 to promote usage of a cryptic branch point and aberrant 3' splice site within intron 12 resulting in the generation of this isoform. These data provide a mechanistic basis for the frequent cooccurrence of SF3B1 mutations as well as new insights into the pathogenesis of myeloid leukemias harboring inv(3)/t(3;3).
© 2022 by The American Society of Hematology.
Figures
Comment in
-
Mutant SF3B1 splices a more leukemogenic EVI1.Blood. 2022 Aug 25;140(8):800-801. doi: 10.1182/blood.2022017380. Blood. 2022. PMID: 36006671 No abstract available.
References
-
- Sun J, Konoplev SN, Wang X, et al. . De novo acute myeloid leukemia with inv(3)(q21q26.2) or t(3;3)(q21;q26.2): a clinicopathologic and cytogenetic study of an entity recently added to the WHO classification. Mod Pathol. 2011;24(3): 384-389. - PubMed
-
- Lugthart S, Gröschel S, Beverloo HB, et al. . Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol. 2010;28(24):3890-3898. - PubMed
-
- Cui W, Sun J, Cotta CV, Medeiros LJ, Lin P. Myelodysplastic syndrome with inv(3)(q21q26.2) or t(3;3)(q21;q26.2) has a high risk for progression to acute myeloid leukemia. Am J Clin Pathol. 2011;136(2): 282-288. - PubMed
-
- Rogers HJ, Vardiman JW, Anastasi J, et al. . Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv(3)(q21q26.2)/t(3;3)(q21;q26.2): a Bone Marrow Pathology Group study. Haematologica. 2014;99(5):821-829. - PMC - PubMed
-
- Gröschel S, Lugthart S, Schlenk RF, et al. . High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28(12):2101-2107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

