Liquid Phase Partitioning in Virus Replication: Observations and Opportunities
- PMID: 35709511
- PMCID: PMC11331907
- DOI: 10.1146/annurev-virology-093020-013659
Liquid Phase Partitioning in Virus Replication: Observations and Opportunities
Abstract
Viruses frequently carry out replication in specialized compartments within cells. The effect of these structures on virus replication is poorly understood. Recent research supports phase separation as a foundational principle for organization of cellular components with the potential to influence viral replication. In this review, phase separation is described in the context of formation of viral replication centers, with an emphasis on the nonsegmented negative-strand RNA viruses. Consideration is given to the interplay between phase separation and the critical processes of viral transcription and genome replication, and the role of these regions in pathogen-host interactions is discussed. Finally, critical questions that must be addressed to fully understand how phase separation influences viral replication and the viral life cycle are presented, along with information about new approaches that could be used to make important breakthroughs in this emerging field.
Keywords: NNSVs; host-pathogen interactions; inclusion bodies; phase separation; viral life cycle; virus transcription and replication.
Figures
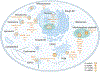

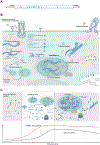
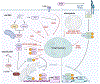
References
-
- Paul D, Bartenschlager R. 2015. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annual Review of Virology 2: 289–310 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

