Accelerators for Classical Molecular Dynamics Simulations of Biomolecules
- PMID: 35710099
- PMCID: PMC9281402
- DOI: 10.1021/acs.jctc.1c01214
Accelerators for Classical Molecular Dynamics Simulations of Biomolecules
Abstract
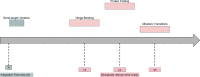
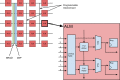
Atomistic Molecular Dynamics (MD) simulations provide researchers the ability to model biomolecular structures such as proteins and their interactions with drug-like small molecules with greater spatiotemporal resolution than is otherwise possible using experimental methods. MD simulations are notoriously expensive computational endeavors that have traditionally required massive investment in specialized hardware to access biologically relevant spatiotemporal scales. Our goal is to summarize the fundamental algorithms that are employed in the literature to then highlight the challenges that have affected accelerator implementations in practice. We consider three broad categories of accelerators: Graphics Processing Units (GPUs), Field-Programmable Gate Arrays (FPGAs), and Application Specific Integrated Circuits (ASICs). These categories are comparatively studied to facilitate discussion of their relative trade-offs and to gain context for the current state of the art. We conclude by providing insights into the potential of emerging hardware platforms and algorithms for MD.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (accessed 10-11-2021).
-
- Schneider P.; Walters W. P.; Plowright A. T.; Sieroka N.; Listgarten J.; Goodnow R. A. Jr; Fisher J.; Jansen J. M.; Duca J. S.; Rush T. S.; Zentgraf M.; Hill J. E.; Krutoholow E.; Kohler M.; Blaney J.; Funatsu K.; Luebkemann C.; Schneider G. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discovery 2020, 19, 353–364. 10.1038/s41573-019-0050-3. - DOI - PubMed

