Recurrent chromosomal translocations in sarcomas create a megacomplex that mislocalizes NuA4/TIP60 to Polycomb target loci
- PMID: 35710139
- PMCID: PMC9296003
- DOI: 10.1101/gad.348982.121
Recurrent chromosomal translocations in sarcomas create a megacomplex that mislocalizes NuA4/TIP60 to Polycomb target loci
Abstract
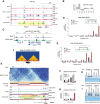
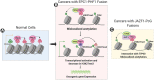
Chromosomal translocations frequently promote carcinogenesis by producing gain-of-function fusion proteins. Recent studies have identified highly recurrent chromosomal translocations in patients with endometrial stromal sarcomas (ESSs) and ossifying fibromyxoid tumors (OFMTs), leading to an in-frame fusion of PHF1 (PCL1) to six different subunits of the NuA4/TIP60 complex. While NuA4/TIP60 is a coactivator that acetylates chromatin and loads the H2A.Z histone variant, PHF1 is part of the Polycomb repressive complex 2 (PRC2) linked to transcriptional repression of key developmental genes through methylation of histone H3 on lysine 27. In this study, we characterize the fusion protein produced by the EPC1-PHF1 translocation. The chimeric protein assembles a megacomplex harboring both NuA4/TIP60 and PRC2 activities and leads to mislocalization of chromatin marks in the genome, in particular over an entire topologically associating domain including part of the HOXD cluster. This is linked to aberrant gene expression-most notably increased expression of PRC2 target genes. Furthermore, we show that JAZF1-implicated with a PRC2 component in the most frequent translocation in ESSs, JAZF1-SUZ12-is a potent transcription activator that physically associates with NuA4/TIP60, its fusion creating outcomes similar to those of EPC1-PHF1 Importantly, the specific increased expression of PRC2 targets/HOX genes was also confirmed with ESS patient samples. Altogether, these results indicate that most chromosomal translocations linked to these sarcomas use the same molecular oncogenic mechanism through a physical merge of NuA4/TIP60 and PRC2 complexes, leading to mislocalization of histone marks and aberrant Polycomb target gene expression.
Keywords: EPC1; H3K27 methylation; H4 acetylation; HOXD; JAZF1; NuA4; PHF1; PRC2; SUZ12; TIP60.
© 2022 Sudarshan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.
-
- Antonescu CR, Sung YS, Chen CL, Zhang L, Chen HW, Singer S, Agaram NP, Sboner A, Fletcher CD. 2014. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors—molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer 53: 183–193. 10.1002/gcc.22132 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
