Empowering the discovery of novel target-disease associations via machine learning approaches in the open targets platform
- PMID: 35710324
- PMCID: PMC9202116
- DOI: 10.1186/s12859-022-04753-4
Empowering the discovery of novel target-disease associations via machine learning approaches in the open targets platform
Abstract
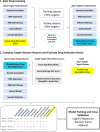
Background: The Open Targets (OT) Platform integrates a wide range of data sources on target-disease associations to facilitate identification of potential therapeutic drug targets to treat human diseases. However, due to the complexity that targets are usually functionally pleiotropic and efficacious for multiple indications, challenges in identifying novel target to indication associations remain. Specifically, persistent need exists for new methods for integration of novel target-disease association evidence and biological knowledge bases via advanced computational methods. These offer promise for increasing power for identification of the most promising target-disease pairs for therapeutic development. Here we introduce a novel approach by integrating additional target-disease features with machine learning models to further uncover druggable disease to target indications.
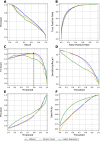
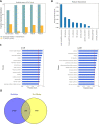
Results: We derived novel target-disease associations as supplemental features to OT platform-based associations using three data sources: (1) target tissue specificity from GTEx expression profiles; (2) target semantic similarities based on gene ontology; and (3) functional interactions among targets by embedding them from protein-protein interaction (PPI) networks. Machine learning models were applied to evaluate feature importance and performance benchmarks for predicting targets with known drug indications. The evaluation results show the newly integrated features demonstrate higher importance than current features in OT. In addition, these also show superior performance over association benchmarks and may support discovery of novel therapeutic indications for highly pursued targets.
Conclusion: Our newly generated features can be used to represent additional underlying biological relatedness among targets and diseases to further empower improved performance for predicting novel indications for drug targets through advanced machine learning models. The proposed methodology enables a powerful new approach for systematic evaluation of drug targets with novel indications.
Keywords: Data Integration; Drug discovery; Drug repurposing; Feature engineering; Machine learning; Open targets; Target indication expansion; XGBoost.
© 2022. The Author(s).
Conflict of interest statement
YH, KK, DKR, CZ, and ET are employees of Sanofi and may hold shares and/or stock options in the company.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

