Machine learning approaches to the human metabolome in sepsis identify metabolic links with survival
- PMID: 35710638
- PMCID: PMC9203139
- DOI: 10.1186/s40635-022-00445-8
Machine learning approaches to the human metabolome in sepsis identify metabolic links with survival
Abstract
Background: Metabolic predictors and potential mediators of survival in sepsis have been incompletely characterized. We examined whether machine learning (ML) tools applied to the human plasma metabolome could consistently identify and prioritize metabolites implicated in sepsis survivorship, and whether these methods improved upon conventional statistical approaches.
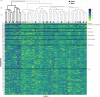
Methods: Plasma gas chromatography-liquid chromatography mass spectrometry quantified 411 metabolites measured ≤ 72 h of ICU admission in 60 patients with sepsis at a single center (Brigham and Women's Hospital, Boston, USA). Seven ML approaches were trained to differentiate survivors from non-survivors. Model performance predicting 28 day mortality was assessed through internal cross-validation, and innate top-feature (metabolite) selection and rankings were compared across the 7 ML approaches and with conventional statistical methods (logistic regression). Metabolites were consensus ranked by a summary, ensemble ML ranking procedure weighing their contribution to mortality risk prediction across multiple ML models.
Results: Median (IQR) patient age was 58 (47, 62) years, 45% were women, and median (IQR) SOFA score was 9 (6, 12). Mortality at 28 days was 42%. The models' specificity ranged from 0.619 to 0.821. Partial least squares regression-discriminant analysis and nearest shrunken centroids prioritized the greatest number of metabolites identified by at least one other method. Penalized logistic regression demonstrated top-feature results that were consistent with many ML methods. Across the plasma metabolome, the 13 metabolites with the strongest linkage to mortality defined through an ensemble ML importance score included lactate, bilirubin, kynurenine, glycochenodeoxycholate, phenylalanine, and others. Four of these top 13 metabolites (3-hydroxyisobutyrate, indoleacetate, fucose, and glycolithocholate sulfate) have not been previously associated with sepsis survival. Many of the prioritized metabolites are constituents of the tryptophan, pyruvate, phenylalanine, pentose phosphate, and bile acid pathways.
Conclusions: We identified metabolites linked with sepsis survival, some confirming prior observations, and others representing new associations. The application of ensemble ML feature-ranking tools to metabolomic data may represent a promising statistical platform to support biologic target discovery.
Keywords: Artificial intelligence; Machine learning; Metabolism; Metabolomics; Sepsis.
© 2022. The Author(s).
Conflict of interest statement
RMB takes part in advisory boards for Merck and Genentech; all remaining authors have no relevant financial disclosures.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

