CRISPR/Cas9 editing of the MYO7A gene in rhesus macaque embryos to generate a primate model of Usher syndrome type 1B
- PMID: 35710827
- PMCID: PMC9203743
- DOI: 10.1038/s41598-022-13689-x
CRISPR/Cas9 editing of the MYO7A gene in rhesus macaque embryos to generate a primate model of Usher syndrome type 1B
Abstract
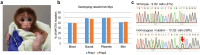
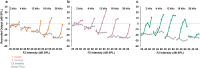
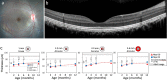
Mutations in the MYO7A gene lead to Usher syndrome type 1B (USH1B), a disease characterized by congenital deafness, vision loss, and balance impairment. To create a nonhuman primate (NHP) USH1B model, CRISPR/Cas9 was used to disrupt MYO7A in rhesus macaque zygotes. The targeting efficiency of Cas9 mRNA and hybridized crRNA-tracrRNA (hyb-gRNA) was compared to Cas9 nuclease (Nuc) protein and synthetic single guide (sg)RNAs. Nuc/sgRNA injection led to higher editing efficiencies relative to mRNA/hyb-gRNAs. Mutations were assessed by preimplantation genetic testing (PGT) and those with the desired mutations were transferred into surrogates. A pregnancy was established from an embryo where 92.1% of the PGT sequencing reads possessed a single G insertion that leads to a premature stop codon. Analysis of single peripheral blood leukocytes from the infant revealed that half the cells possessed the homozygous single base insertion and the remaining cells had the wild-type MYO7A sequence. The infant showed sensitive auditory thresholds beginning at 3 months. Although further optimization is needed, our studies demonstrate that it is feasible to use CRISPR technologies for creating NHP models of human diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

