The cohesin acetylation cycle controls chromatin loop length through a PDS5A brake mechanism
- PMID: 35710836
- PMCID: PMC9205776
- DOI: 10.1038/s41594-022-00773-z
The cohesin acetylation cycle controls chromatin loop length through a PDS5A brake mechanism
Abstract
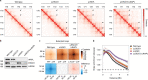
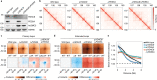
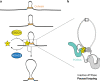
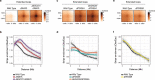
Cohesin structures the genome through the formation of chromatin loops and by holding together the sister chromatids. The acetylation of cohesin's SMC3 subunit is a dynamic process that involves the acetyltransferase ESCO1 and deacetylase HDAC8. Here we show that this cohesin acetylation cycle controls the three-dimensional genome in human cells. ESCO1 restricts the length of chromatin loops, and of architectural stripes emanating from CTCF sites. HDAC8 conversely promotes the extension of such loops and stripes. This role in controlling loop length turns out to be distinct from the canonical role of cohesin acetylation that protects against WAPL-mediated DNA release. We reveal that acetylation controls the interaction of cohesin with PDS5A to restrict chromatin loop length. Our data support a model in which this PDS5A-bound state acts as a brake that enables the pausing and restart of loop enlargement. The cohesin acetylation cycle hereby provides punctuation in the process of genome folding.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

