Molecular and cellular immune features of aged patients with severe COVID-19 pneumonia
- PMID: 35710943
- PMCID: PMC9203559
- DOI: 10.1038/s42003-022-03537-z
Molecular and cellular immune features of aged patients with severe COVID-19 pneumonia
Abstract
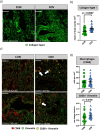
Aging is a major risk factor for developing severe COVID-19, but few detailed data are available concerning immunological changes after infection in aged individuals. Here we describe main immune characteristics in 31 patients with severe SARS-CoV-2 infection who were >70 years old, compared to 33 subjects <60 years of age. Differences in plasma levels of 62 cytokines, landscape of peripheral blood mononuclear cells, T cell repertoire, transcriptome of central memory CD4+ T cells, specific antibodies are reported along with features of lung macrophages. Elderly subjects have higher levels of pro-inflammatory cytokines, more circulating plasmablasts, reduced plasmatic level of anti-S and anti-RBD IgG3 antibodies, lower proportions of central memory CD4+ T cells, more immature monocytes and CD56+ pro-inflammatory monocytes, lower percentages of circulating follicular helper T cells (cTfh), antigen-specific cTfh cells with a less activated transcriptomic profile, lung resident activated macrophages that promote collagen deposition and fibrosis. Our study underlines the importance of inflammation in the response to SARS-CoV-2 and suggests that inflammaging, coupled with the inability to mount a proper anti-viral response, could exacerbate disease severity and the worst clinical outcome in old patients.
© 2022. The Author(s).
Conflict of interest statement
AQ, CP, GA, SD, DL, and JN are employers of Fluidigm Corporation. All other authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

