Rapid evaluation of COVID-19 vaccine effectiveness against symptomatic infection with SARS-CoV-2 variants by analysis of genetic distance
- PMID: 35710987
- PMCID: PMC9388371
- DOI: 10.1038/s41591-022-01877-1
Rapid evaluation of COVID-19 vaccine effectiveness against symptomatic infection with SARS-CoV-2 variants by analysis of genetic distance
Abstract
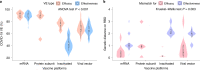
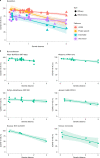
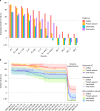
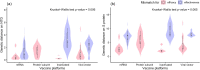
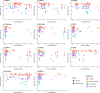
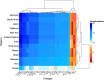
Timely evaluation of the protective effects of Coronavirus Disease 2019 (COVID-19) vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern is urgently needed to inform pandemic control planning. Based on 78 vaccine efficacy or effectiveness (VE) data from 49 studies and 1,984,241 SARS-CoV-2 sequences collected from 31 regions, we analyzed the relationship between genetic distance (GD) of circulating viruses against the vaccine strain and VE against symptomatic infection. We found that the GD of the receptor-binding domain of the SARS-CoV-2 spike protein is highly predictive of vaccine protection and accounted for 86.3% (P = 0.038) of the VE change in a vaccine platform-based mixed-effects model and 87.9% (P = 0.006) in a manufacturer-based model. We applied the VE-GD model to predict protection mediated by existing vaccines against new genetic variants and validated the results by published real-world and clinical trial data, finding high concordance of predicted VE with observed VE. We estimated the VE against the Delta variant to be 82.8% (95% prediction interval: 68.7-96.0) using the mRNA vaccine platform, closely matching the reported VE of 83.0% from an observational study. Among the four sublineages of Omicron, the predicted VE varied between 11.9% and 33.3%, with the highest VE predicted against BA.1 and the lowest against BA.2, using the mRNA vaccine platform. The VE-GD framework enables predictions of vaccine protection in real time and offers a rapid evaluation method against novel variants that may inform vaccine deployment and public health responses.
© 2022. The Author(s).
Conflict of interest statement
M.H.W. and B.C.Y.Z. are shareholders of Beth Bioinformatics Co., Ltd. B.C.Y.Z. is a shareholder of Health View Bioanalytics Ltd. S.Y.C. and J.L. are employees of Beth Bioinformatics Co., Ltd. All other authors declare no competing interests.
Figures




References
-
- Basta, N. E. & Moodie, E. M. M., on behalf of the VIPER (Vaccines, Infectious disease Prevention, and Epidemiology Research) Group COVID-19 Vaccine Development and Approvals Tracker Team. COVID-19 Vaccine Tracker. https://covid19.trackvaccines.org/
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

