A single-cell transcriptomic atlas tracking the neural basis of division of labour in an ant superorganism
- PMID: 35711063
- PMCID: PMC9349048
- DOI: 10.1038/s41559-022-01784-1
A single-cell transcriptomic atlas tracking the neural basis of division of labour in an ant superorganism
Abstract
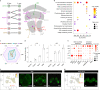
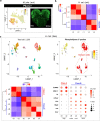
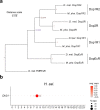
Ant colonies with permanent division of labour between castes and highly distinct roles of the sexes have been conceptualized to be superorganisms, but the cellular and molecular mechanisms that mediate caste/sex-specific behavioural specialization have remained obscure. Here we characterized the brain cell repertoire of queens, gynes (virgin queens), workers and males of Monomorium pharaonis by obtaining 206,367 single-nucleus transcriptomes. In contrast to Drosophila, the mushroom body Kenyon cells are abundant in ants and display a high diversity with most subtypes being enriched in worker brains, the evolutionarily derived caste. Male brains are as specialized as worker brains but with opposite trends in cell composition with higher abundances of all optic lobe neuronal subtypes, while the composition of gyne and queen brains remained generalized, reminiscent of solitary ancestors. Role differentiation from virgin gynes to inseminated queens induces abundance changes in roughly 35% of cell types, indicating active neurogenesis and/or programmed cell death during this transition. We also identified insemination-induced cell changes probably associated with the longevity and fecundity of the reproductive caste, including increases of ensheathing glia and a population of dopamine-regulated Dh31-expressing neurons. We conclude that permanent caste differentiation and extreme sex-differentiation induced major changes in the neural circuitry of ants.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
Complementary brains.Nat Ecol Evol. 2022 Aug;6(8):1060-1061. doi: 10.1038/s41559-022-01805-z. Nat Ecol Evol. 2022. PMID: 35711064 No abstract available.
References
-
- Wheeler, W. M. Ants: Their Structure, Development and Behavior (Columbia Univ. Press, 1910).
-
- Wheeler WM. The ant‐colony as an organism. J. Morphol. 1911;22:307–325. doi: 10.1002/jmor.1050220206. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

