Encoding with a fluorescence-activating and absorption-shifting tag generates living bacterial probes for mammalian microbiota imaging
- PMID: 35711290
- PMCID: PMC9194656
- DOI: 10.1016/j.mtbio.2022.100311
Encoding with a fluorescence-activating and absorption-shifting tag generates living bacterial probes for mammalian microbiota imaging
Abstract
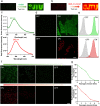
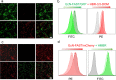
The mammalian microbiota plays essential roles in health. A primary determinant to understand the interaction with the host is the distribution and viability of its key microorganisms. Here, a strategy of encoding with a fluorescence-activating and absorption-shifting tag (FAST) is reported to prepare living bacterial probes for real-time dynamic, dual-modal, and molecular oxygen-independent imaging of the host microbiota. Carrying FAST endows bacteria with rapid on-demand turn on-off fluorescence by adding or removal of corresponding fluorogens. Encoded bacteria are able to reversibly switch emission bands for dual-color fluorescence imaging via fluorogen exchange. Due to molecular oxygen-independent emission of FAST, encoded bacteria can emit fluorescence under anaerobic environments including the gut and tumor. These living probes demonstrate the applicability to quantify the vitality of bacteria transplanted to the gut microbiota. This work proposes a unique fluorescence probe for investigating the dynamics of the host microbiota.
Keywords: Absorption-shifting; Bacteria; Fluorescence-activating; Gut microbiota; Imaging; Tumor.
© 2022 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Heintz-Buschart A., Wilmes P. Human gut microbiome: function matters. Trends Microbiol. 2018;26:563–574. - PubMed
-
- Budden K.F., Gellatly S.L., Wood D.L., Cooper M.A., Morrison M., Hugenholtz P., Hansbro P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017;15:55–63. - PubMed
-
- Helmink B.A., Khan M.A.W., Hermann A., Gopalakrishnan V., Wargo J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019;25:377–388. - PubMed
LinkOut - more resources
Full Text Sources

