Population dynamics and genetic connectivity in recent chimpanzee history
- PMID: 35711737
- PMCID: PMC9188271
- DOI: 10.1016/j.xgen.2022.100133
Population dynamics and genetic connectivity in recent chimpanzee history
Abstract
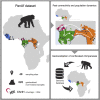
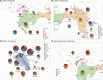
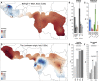
Knowledge on the population history of endangered species is critical for conservation, but whole-genome data on chimpanzees (Pan troglodytes) is geographically sparse. Here, we produced the first non-invasive geolocalized catalog of genomic diversity by capturing chromosome 21 from 828 non-invasive samples collected at 48 sampling sites across Africa. The four recognized subspecies show clear genetic differentiation correlating with known barriers, while previously undescribed genetic exchange suggests that these have been permeable on a local scale. We obtained a detailed reconstruction of population stratification and fine-scale patterns of isolation, migration, and connectivity, including a comprehensive picture of admixture with bonobos (Pan paniscus). Unlike humans, chimpanzees did not experience extended episodes of long-distance migrations, which might have limited cultural transmission. Finally, based on local rare variation, we implement a fine-grained geolocalization approach demonstrating improved precision in determining the origin of confiscated chimpanzees.
Keywords: chimpanzee; chimpanzee demography; conservation genomics; fecal samples; geolocalization; hybridization capture; non-invasive samples; population dynamics; population genetics.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Lobon I., Tucci S., De Manuel M., Ghirotto S., Benazzo A., Prado-Martinez J., Lorente-Galdos B., Nam K., Dabad M., Hernandez-Rodriguez J., et al. Demographic history of the genus Pan inferred from whole mitochondrial genome reconstructions. Genome Biol. Evol. 2016;8:2020–2030. doi: 10.1093/gbe/evw124. - DOI - PMC - PubMed
-
- Hallast P., Maisano Delser P., Batini C., Zadik D., Rocchi M., Schempp W., Tyler-Smith C., Jobling M.A. Great ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal. Genome Res. 2016;26:427–439. doi: 10.1101/gr.198754.115. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

