StarGazer: A Hybrid Intelligence Platform for Drug Target Prioritization and Digital Drug Repositioning Using Streamlit
- PMID: 35711912
- PMCID: PMC9197487
- DOI: 10.3389/fgene.2022.868015
StarGazer: A Hybrid Intelligence Platform for Drug Target Prioritization and Digital Drug Repositioning Using Streamlit
Abstract
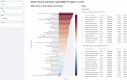
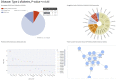
Target prioritization is essential for drug discovery and repositioning. Applying computational methods to analyze and process multi-omics data to find new drug targets is a practical approach for achieving this. Despite an increasing number of methods for generating datasets such as genomics, phenomics, and proteomics, attempts to integrate and mine such datasets remain limited in scope. Developing hybrid intelligence solutions that combine human intelligence in the scientific domain and disease biology with the ability to mine multiple databases simultaneously may help augment drug target discovery and identify novel drug-indication associations. We believe that integrating different data sources using a singular numerical scoring system in a hybrid intelligent framework could help to bridge these different omics layers and facilitate rapid drug target prioritization for studies in drug discovery, development or repositioning. Herein, we describe our prototype of the StarGazer pipeline which combines multi-source, multi-omics data with a novel target prioritization scoring system in an interactive Python-based Streamlit dashboard. StarGazer displays target prioritization scores for genes associated with 1844 phenotypic traits, and is available via https://github.com/AstraZeneca/StarGazer.
Keywords: data integration; drug discovery; hybrid intelligence; multi-omics; repositioning; stargazer; streamlit; target prioritization.
Copyright © 2022 Lee, Lin, Prokop, Gopalakrishnan, Hanna, Papa, Freeman, Patel, Yu, Huhn, Sheikh, Tan, Sellman, Cohen, Mangion, Khan, Gusev and Shameer.
Conflict of interest statement
Authors CL, AP, VG, RNH, EP, AF, SP, WY, MH, A-SS, KT, BS, TC, JM, FMK, and KS are or were employed by AstraZeneca. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
integRATE: a desirability-based data integration framework for the prioritization of candidate genes across heterogeneous omics and its application to preterm birth.BMC Med Genomics. 2018 Nov 19;11(1):107. doi: 10.1186/s12920-018-0426-y. BMC Med Genomics. 2018. PMID: 30453955 Free PMC article.
-
COMO: a pipeline for multi-omics data integration in metabolic modeling and drug discovery.Brief Bioinform. 2023 Sep 22;24(6):bbad387. doi: 10.1093/bib/bbad387. Brief Bioinform. 2023. PMID: 37930022 Free PMC article.
-
Computational Study of Drugs by Integrating Omics Data with Kernel Methods.Mol Inform. 2013 Dec;32(11-12):930-41. doi: 10.1002/minf.201300090. Epub 2013 Dec 11. Mol Inform. 2013. PMID: 27481139 Review.
-
PS4DR: a multimodal workflow for identification and prioritization of drugs based on pathway signatures.BMC Bioinformatics. 2020 Jun 5;21(1):231. doi: 10.1186/s12859-020-03568-5. BMC Bioinformatics. 2020. PMID: 32503412 Free PMC article.
-
A primer on applying AI synergistically with domain expertise to oncology.Biochim Biophys Acta Rev Cancer. 2021 Aug;1876(1):188548. doi: 10.1016/j.bbcan.2021.188548. Epub 2021 Apr 24. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33901609 Review.
Cited by
-
SciLinker: a large-scale text mining framework for mapping associations among biological entities.Front Artif Intell. 2025 Mar 19;8:1528562. doi: 10.3389/frai.2025.1528562. eCollection 2025. Front Artif Intell. 2025. PMID: 40212086 Free PMC article.
-
Stmol: A component for building interactive molecular visualizations within streamlit web-applications.Front Mol Biosci. 2022 Sep 23;9:990846. doi: 10.3389/fmolb.2022.990846. eCollection 2022. Front Mol Biosci. 2022. PMID: 36213112 Free PMC article.
-
StreamChol: a web-based application for predicting cholestasis.J Cheminform. 2025 Jan 21;17(1):9. doi: 10.1186/s13321-024-00943-9. J Cheminform. 2025. PMID: 39838478 Free PMC article.
-
CPGminer: An Interactive Dashboard to Explore the Genomic Features and Taxonomy of Complete Prokaryotic Genomes.Microorganisms. 2023 Oct 13;11(10):2556. doi: 10.3390/microorganisms11102556. Microorganisms. 2023. PMID: 37894214 Free PMC article.
-
The landscape of the methodology in drug repurposing using human genomic data: a systematic review.Brief Bioinform. 2024 Jan 22;25(2):bbad527. doi: 10.1093/bib/bbad527. Brief Bioinform. 2024. PMID: 38279645 Free PMC article.
References
-
- Akata Z., Eiben G., Fokkens A., Grossi D., Hindriks K., Hoos H., et al. (2020). A Research Agenda for Hybrid Intelligence: Augmenting Human Intellect with Collaborative, Adaptive, Responsible, and Explainable Artificial Intelligence. Computer 53, 18–28. 10.1109/mc.2020.2996587 - DOI
-
- Armstrong J. F., Faccenda E., Harding S. D., Pawson A. J., Southan C., Sharman J. L., et al. (2019). The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: Extending Immunopharmacology Content and Introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res. 48, D1006–D1021. 10.1093/nar/gkz951 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

