Dynamic Meta-data Network Sparse PCA for Cancer Subtype Biomarker Screening
- PMID: 35711917
- PMCID: PMC9197542
- DOI: 10.3389/fgene.2022.869906
Dynamic Meta-data Network Sparse PCA for Cancer Subtype Biomarker Screening
Abstract
Previous research shows that each type of cancer can be divided into multiple subtypes, which is one of the key reasons that make cancer difficult to cure. Under these circumstances, finding a new target gene of cancer subtypes has great significance on developing new anti-cancer drugs and personalized treatment. Due to the fact that gene expression data sets of cancer are usually high-dimensional and with high noise and have multiple potential subtypes' information, many sparse principal component analysis (sparse PCA) methods have been used to identify cancer subtype biomarkers and subtype clusters. However, the existing sparse PCA methods have not used the known cancer subtype information as prior knowledge, and their results are greatly affected by the quality of the samples. Therefore, we propose the Dynamic Metadata Edge-group Sparse PCA (DM-ESPCA) model, which combines the idea of meta-learning to solve the problem of sample quality and uses the known cancer subtype information as prior knowledge to capture some gene modules with better biological interpretations. The experiment results on the three biological data sets showed that the DM-ESPCA model can find potential target gene probes with richer biological information to the cancer subtypes. Moreover, the results of clustering and machine learning classification models based on the target genes screened by the DM-ESPCA model can be improved by up to 22-23% of accuracies compared with the existing sparse PCA methods. We also proved that the result of the DM-ESPCA model is better than those of the four classic supervised machine learning models in the task of classification of cancer subtypes.
Keywords: Cancer subtype; DM-ESPCA model; biomarkers; dynamic network; meta-data; sparse PCA.
Copyright © 2022 Miao, Dong, Liu, Lo, Mei, Dang, Cai, Li, Yang, Xie and Liang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


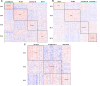
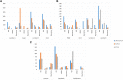



References
-
- Carlson M., Falcon S., Pages H., Li N. (2016). hgu133plus2. Db: Affymetrix Human Genome U133 Plus 2.0 Array Annotation Data (Chip Hgu133plus2). R. Package Version 3.
LinkOut - more resources
Full Text Sources

