NNMT: Mean-Field Based Analysis Tools for Neuronal Network Models
- PMID: 35712677
- PMCID: PMC9196133
- DOI: 10.3389/fninf.2022.835657
NNMT: Mean-Field Based Analysis Tools for Neuronal Network Models
Abstract
Mean-field theory of neuronal networks has led to numerous advances in our analytical and intuitive understanding of their dynamics during the past decades. In order to make mean-field based analysis tools more accessible, we implemented an extensible, easy-to-use open-source Python toolbox that collects a variety of mean-field methods for the leaky integrate-and-fire neuron model. The Neuronal Network Mean-field Toolbox (NNMT) in its current state allows for estimating properties of large neuronal networks, such as firing rates, power spectra, and dynamical stability in mean-field and linear response approximation, without running simulations. In this article, we describe how the toolbox is implemented, show how it is used to reproduce results of previous studies, and discuss different use-cases, such as parameter space explorations, or mapping different network models. Although the initial version of the toolbox focuses on methods for leaky integrate-and-fire neurons, its structure is designed to be open and extensible. It aims to provide a platform for collecting analytical methods for neuronal network model analysis, such that the neuroscientific community can take maximal advantage of them.
Keywords: (hybrid) modeling; (spiking) neuronal network; computational neuroscience; integrate-and-fire neuron; mean-field theory; open-source software; parameter space exploration; python.
Copyright © 2022 Layer, Senk, Essink, van Meegen, Bos and Helias.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






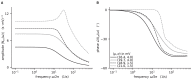


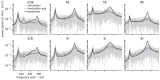

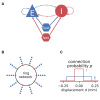

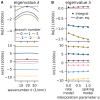
References
-
- Abramowitz M., Stegun I. A. (1974). Handbook of Mathematical Functions: With Formulas, Graphs, and Mathematical Tables (New York: Dover Publications; ).
LinkOut - more resources
Full Text Sources
Miscellaneous

