NetRAX: accurate and fast maximum likelihood phylogenetic network inference
- PMID: 35713506
- PMCID: PMC9344847
- DOI: 10.1093/bioinformatics/btac396
NetRAX: accurate and fast maximum likelihood phylogenetic network inference
Abstract
Motivation: Phylogenetic networks can represent non-treelike evolutionary scenarios. Current, actively developed approaches for phylogenetic network inference jointly account for non-treelike evolution and incomplete lineage sorting (ILS). Unfortunately, this induces a very high computational complexity and current tools can only analyze small datasets.
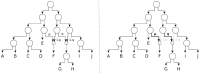
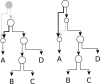
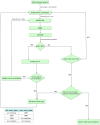
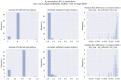
Results: We present NetRAX, a tool for maximum likelihood (ML) inference of phylogenetic networks in the absence of ILS. Our tool leverages state-of-the-art methods for efficiently computing the phylogenetic likelihood function on trees, and extends them to phylogenetic networks via the notion of 'displayed trees'. NetRAX can infer ML phylogenetic networks from partitioned multiple sequence alignments and returns the inferred networks in Extended Newick format. On simulated data, our results show a very low relative difference in Bayesian Information Criterion (BIC) score and a near-zero unrooted softwired cluster distance to the true, simulated networks. With NetRAX, a network inference on a partitioned alignment with 8000 sites, 30 taxa and 3 reticulations completes within a few minutes on a standard laptop.
Availability and implementation: Our implementation is available under the GNU General Public License v3.0 at https://github.com/lutteropp/NetRAX.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures


References
-
- Allen-Savietta C. (2020) Estimating Phylogenetic Networks from Concatenated Sequence Alignments. The University of Wisconsin-Madison, Madison, Wisconsin, USA.
-
- Ané C. (2021) Phylonetworks Users Google Group Discussion. https://groups.google.com/g/phylonetworks-users/c/KCu45cDRy_Q/m/RLpaZJaj... (14 August 2021, date last accessed).
-
- Blair C., Ané C. (2019) Phylogenetic trees and networks can serve as powerful and complementary approaches for analysis of genomic data. Syst. Biol., 69, 593–601. - PubMed
-
- Burbrink F.T., Gehara M. (2018) The biogeography of deep time phylogenetic reticulation. Syst. Biol., 67, 743–755. - PubMed
-
- Cao Z. et al. (2019) Practical aspects of phylogenetic network analysis using phylonet. BioRxiv, page 746362.

