Dominant-acting CSF1R variants cause microglial depletion and altered astrocytic phenotype in zebrafish and adult-onset leukodystrophy
- PMID: 35713703
- PMCID: PMC9288387
- DOI: 10.1007/s00401-022-02440-5
Dominant-acting CSF1R variants cause microglial depletion and altered astrocytic phenotype in zebrafish and adult-onset leukodystrophy
Abstract
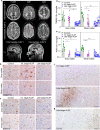
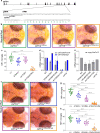
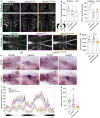
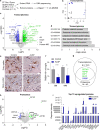
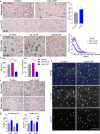
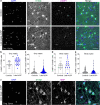
Tissue-resident macrophages of the brain, including microglia, are implicated in the pathogenesis of various CNS disorders and are possible therapeutic targets by their chemical depletion or replenishment by hematopoietic stem cell therapy. Nevertheless, a comprehensive understanding of microglial function and the consequences of microglial depletion in the human brain is lacking. In human disease, heterozygous variants in CSF1R, encoding the Colony-stimulating factor 1 receptor, can lead to adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP) possibly caused by microglial depletion. Here, we investigate the effects of ALSP-causing CSF1R variants on microglia and explore the consequences of microglial depletion in the brain. In intermediate- and late-stage ALSP post-mortem brain, we establish that there is an overall loss of homeostatic microglia and that this is predominantly seen in the white matter. By introducing ALSP-causing missense variants into the zebrafish genomic csf1ra locus, we show that these variants act dominant negatively on the number of microglia in vertebrate brain development. Transcriptomics and proteomics on relatively spared ALSP brain tissue validated a downregulation of microglia-associated genes and revealed elevated astrocytic proteins, possibly suggesting involvement of astrocytes in early pathogenesis. Indeed, neuropathological analysis and in vivo imaging of csf1r zebrafish models showed an astrocytic phenotype associated with enhanced, possibly compensatory, endocytosis. Together, our findings indicate that microglial depletion in zebrafish and human disease, likely as a consequence of dominant-acting pathogenic CSF1R variants, correlates with altered astrocytes. These findings underscore the unique opportunity CSF1R variants provide to gain insight into the roles of microglia in the human brain, and the need to further investigate how microglia, astrocytes, and their interactions contribute to white matter homeostasis.
Keywords: ALSP; Astrocytes; CSF1R; Leukodystrophy; Microglia; Zebrafish models.
© 2022. The Author(s).
Figures


References
-
- Adams SJ, Kirk A, Auer RN. Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP): integrating the literature on hereditary diffuse leukoencephalopathy with spheroids (HDLS) and pigmentary orthochromatic leukodystrophy (POLD) J Clin Neurosci. 2018;48:42–49. doi: 10.1016/j.jocn.2017.10.060. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

